The Research Brief is a short take about interesting academic work.
The big idea
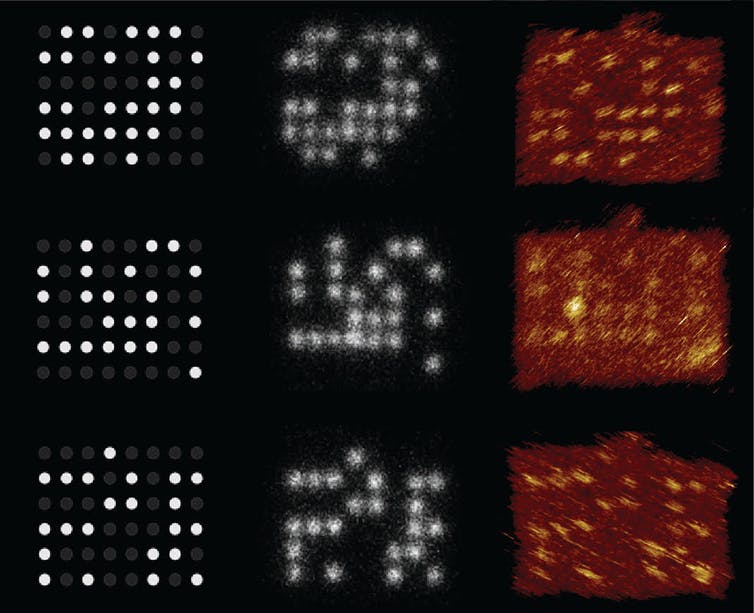
We and our colleagues have developed a way to store data using pegs and pegboards made out of DNA and retrieving the data with a microscope – a molecular version of the Lite-Brite toy. Our prototype stores information in patterns using DNA strands spaced about 10 nanometers apart. Ten nanometers is more than a thousand times smaller than the diameter of a human hair and about 100 times smaller than the diameter of a bacterium.
We tested our digital nucleic acid memory (dNAM) by storing the statement “Data is in our DNA!n.” We described the research in a paper published in the journal Nature Communications on April 22, 2021.
Previous methods for retrieving data in DNA require the DNA to be sequenced. Sequencing is the process of reading the genetic code of strands of DNA. Though it is a powerful tool in medicine and biology, it wasn’t designed with DNA memory in mind.
Our approach uses a microscope to read the data optically. Because the DNA pegs are positioned closer than half the wavelength of visible light, we used super-resolution microscopy, which circumvents the diffraction limit of light. This provides a way to read the encoded data without sequencing the DNA.
Nucleic Acid Memory Institute at Boise State University, CC BY-ND
The patterns of DNA strands – the pegs – light up when fluorescently labeled DNA bind to them. Because the fluorescent strands are short, they rapidly bind and unbind. This causes them to blink, making it easier to separate one peg from another and read the stored information. We use the fluorescent patterns of each pegboard as a code to store chunks of data.
The microscope can image hundreds of thousands of the DNA pegs in a single recording, and our error-correction algorithms ensure we recover all of the data. After accounting for the bits used by the algorithms, our prototype was able to read data at a density of 330 gigabits per square centimeter.
Why it matters
You’re not likely to have a DNA storage device in your phone or computer, at least anytime soon. DNA data storage is promising for archival storage – storing large amounts of information for long periods of time. DNA can store a lot of information in a small space. It would be possible to store every tweet, email, photo, song, movie and book ever created in a volume equivalent to a jewelry box. And data stored in DNA could last for centuries, given that the biomolecule has a half-life of over 500 years.
What other research is being done
Researchers have been developing methods of storing data in DNA for several decades. Those methods involve the design and synthesis of unique strings of information made from the DNA nucleotides adenine (A), thymine (T), cytosine (C) and guanine (G). This information is recovered by reading the strings using sequencing technology.
What’s next
From here, our goal is to increase the amount of data that we can store in dNAM, decrease the amount of time it takes to write and read the data, and use the technique to encrypt data.
[Get our best science, health and technology stories. Sign up for The Conversation’s science newsletter.]