Viruses are the most prevalent biological entities in the world’s oceans and play essential roles in its ecological and biogeochemical balance. Yet, they are the least understood elements of marine life.
By unraveling the entire genome of a certain marine protist that may act as a host for many viruses, an international research team led by scientists from Stony Brook University sets the stage for future investigations of marine protist genomes, marine microbial dynamics and the evolutionary interplay between host organisms and their viruses—work that may open doors to a better understanding of the “invisible” world of marine viruses and offers a key to the ecology and health of oceans worldwide.
The research is published early online in Current Biology.
Food webs of the oceans provide humanity with essential food sources as well as the wonderment of sea creatures from polar bears to penguins. This wellspring of life is supported mainly by microscopic organisms, including the wide presence of viruses. Learning more about the viruses through DNA research and other forms of investigation is essential to scientists’ understanding of the sea. Novel groups of viruses are still being discovered, such as the recently discovered “mirusvirues” featured in a Nature paper earlier this year.
Mirusviruses are large DNA viruses sharing genes with both major realms of viral diversity. They are believed to be ubiquitous in sunlit oceans, important to biodiversity, and not likely harmful to human or aquatic life. The discovery helps scientists better understand the biodiversity of plankton at the surface of the ocean, the importance of these viruses in the ecosystem, and informs on the history of viruses.
The Current Biology study brings more pieces to fit the puzzle of ocean viruses.
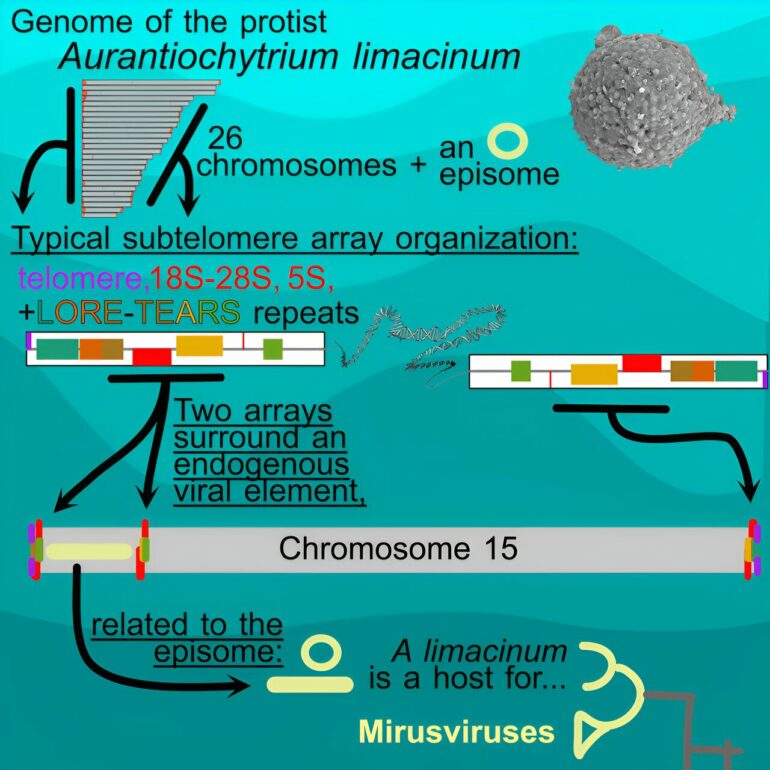
Co-lead authors Jackie Collier, Ph.D., Associate Professor in the School of Marine at Atmospheric Sciences (SoMAS), and Joshua Rest, Ph.D., Associate Professor in the Department of Ecology & Evolution, working with colleagues from Dalhousie University and the Joint Genome Institute, made discoveries about the genome structure of the marine protist Aurantiochytrium limacinum. These findings not only shed light on the unique features of its chromosomes but also uncover the elusive hosts for mirusviruses.
Labyrinthulomycetes, a class to which Aurantiochytrium belongs, have fascinated researchers for their peculiar traits and biotechnological potential (such as production of essential omega-3 fatty acids and carotenoids). Using advanced sequencing technologies, the team was able to completely assemble the genome of Aurantiochytrium.
They discovered two genomic elements with a strong resemblance to mirusviruses. One element is a circular structure present in high copies, while the other integrates within the end of a chromosome. These two states are reminiscent of herpesviruses, infamous for their ability to maintain latent infections in human and other animal hosts: some herpesviruses associated with cancer maintain latent infections by integrating into their host’s genome and others (like chickenpox) maintain latent infections as independent episomes.
The study also dives deep into the structure of Aurantiochytrium’s genome. The team discovered that it possesses a unique configuration at chromosome ends—an unexpected organization of ribosomal RNA genes and long repeats at their subtelomeric regions. The remarkable layout of these sequences may have a pivotal role in chromosome end maintenance, and it is into one such region that a mirusvirus-like genome is integrated.
“Because mirusviruses were found using environmental sequence data, they were identified without known hosts,” says Collier. “Our data show that A. limacinum is probably a natural host of mirusviruses and that a single host has been infected by multiple distinct viruses.”
By completing the entire genome of the protist, the researchers identified the first known host for mirusviruses, and their observations suggest dynamic host-virus genome interactions.
“The discovery of one of the viral sequences at one of the chromosome ends offers intriguing insights into the evolutionary ancestry of these elements,” adds Rest. “This revelation also aligns with growing evidence pointing toward mirusviruses’ significant influence in marine ecosystems.”
More information:
Jackie L. Collier et al, The protist Aurantiochytrium has universal subtelomeric rDNAs and is a host for mirusviruses, Current Biology (2023). DOI: 10.1016/j.cub.2023.10.009
Provided by
Stony Brook University
Citation:
The unraveling of a protist genome could unlock the mystery of marine viruses (2023, October 31)