When Profs. Joel Sussman and Israel Silman were asked to mentor Chinese students online during the COVID-19 pandemic, the last thing they expected to come out of the experience was highly innovative research on protein evolution that could change our understanding of the way new proteins come into being.
“I was skeptical at first—they were all undergraduate students, and communicating via a computer screen didn’t seem too promising,” Sussman recalls. But he and Silman—a Weizmann Institute of Science professorial duo who have hundreds of joint studies on protein structure and function to their credit—agreed to hold tutorials for a team of four students from leading universities across China. The online mentoring was part of the YutChun-Weizmann Program, headed by Weizmann’s Prof. Binghai Yan.
Sussman and Silman told the students to address them by their first names, a practice unheard of in Chinese universities, and encouraged them to develop critical thinking. Still, they expected nothing more than a respectful summary when they asked the students to review an old paper of theirs on protein sequence variations. Instead, the students came back with an in-depth critique, analyzing the study from a contemporary perspective and suggesting that some of its conclusions could be revised using new methods.
Jing Liu, one of the four mentees, says that this was—for her and the other students—a dramatic departure from what they were used to. “In China, a student studying, say, for a master’s degree can’t challenge a Ph.D. candidate or a postdoc—they might get angry or tell the principal investigator,” she explains. She is quick to note, however, that the environment was different at the Guangdong campus of the Technion—Israel Institute of Technology, where she was studying at the time. “I had a supervisor who was willing to listen to me and hold discussions, something that’s hard to find at other universities in China.”
The online tutorials, to the surprise of both sides, soon transformed into discussions. A 2017 study by Czech scientists, which Liu brought to her tutors’ attention, became a major topic of deliberation—one that hinted at an intriguing twist in the history of protein evolution.
Cracks in the folding dogma
As single-celled organisms that once inhabited the Earth evolved into more complex ones, happenstance changes in their DNA, if those changes were beneficial, tended to be conserved, thanks to natural selection, and passed on to higher organisms. That’s why most protein-coding genes in our bodies have equivalents (the scientific term is “homologs”) in many other species along the evolutionary tree, all the way back to yeast or bacteria. As proteins developed, many of them began to fold into complex structures that allowed them to carry out specialized tasks.
Considering that natural selection has been at work for billions of years, it would seem that proteins must have had enough time to evolve all possible useful sequences. In fact, until recently, scientists believed that all existing proteins were born through the refinement of existing sequences, and that truly new proteins had long ceased to appear.
But just over a decade ago, cracks began to develop in this scientific gospel: Evidence emerged that new proteins continue to be born all the time. When scientists began sequencing entire genomes of various organisms, comparisons revealed the presence of genes coding for “newly born” proteins in all species, from bacteria to humans. These proteins are thought to originate in the noncoding regions that make up most of the genome. In this scenario, a stretch of DNA lacking a recipe for proteins acquires, by chance, a set of mutations that convert it into a protein-coding gene.
The Czech study that so intrigued Liu and her tutors had opened an additional crack in the dogma. The Czech researchers had created about 100 sequences of hypothetical proteins by randomly reshuffling existing protein genes like a deck of cards. When they synthesized these “never-born” proteins and tested them in the lab, they discovered that about a third showed signs of folding into compact structures, rather like natural proteins.
“This was totally amazing,” says Sussman. “If someone had asked me before whether a random protein sequence could fold up that way, I’d have said never.”
Silman explains that proteins’ ability to fold is essential to life. Although not all proteins fold, it is folded ones, those with orderly segments, that perform the critical catalytic functions in living organisms. By showing that “never-born” proteins can fold, the Czech study suggested that new proteins can not only be born but may also perform vital new roles.
Born orphans
How does a noncoding DNA segment turn out a “newly-born” protein, and how does this protein become active? What is the time scale for these processes? And can the mechanisms involved be one day exploited in protein design?
To help address these questions, Sussman and Silman decided to conduct what—to the best of their knowledge—became one of the first structural studies of newly born proteins. They launched the project together with Liu, the paper’s first author, and Rongqing Yuan, then a student at Tsinghua University in Beijing. The four met online for a year and a half before completing the research, which was published recently in the journal Proteins: Structure, Function, and Bioinformatics. The other two students, Wei Shao and Jitong Wang, took part in the initial stages of the project; they dropped out at the end of the scheduled tutorial but are co-authors on the published paper.
The team explored the folding potential of “newly-born” proteins with the help of artificial intelligence (AI) tools that—in the past few years—have revolutionized the study of protein structures. These algorithms can, in most cases, now reliably predict a protein’s 3D structure based on its amino acid sequence alone, bypassing the need to grow protein crystals and determine their structures experimentally.
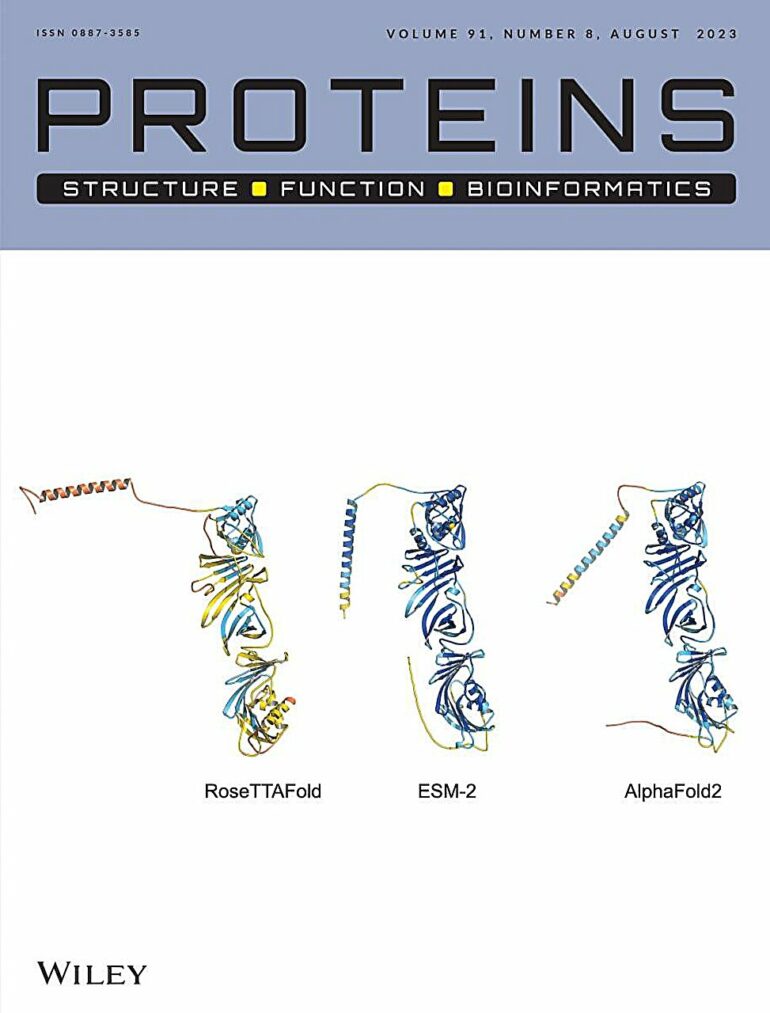
One of the major challenges faced by the team was that these prediction algorithms work best when the protein of interest has numerous homologs (equivalents from other species), whereas “newly-born” proteins, by definition, exist in only one or a handful of species. As they have no evolutionary parents, they are sometimes referred to as orphan proteins (or near-orphans, if they exist in just a few, related species). It took the expertise of the team to apply AI tools to homolog-less orphan proteins successfully. To increase the chances of obtaining trustworthy results, the scientists used three different AI algorithms—AlphaFold2, RoseTTAFold and ESMFold—and compared their findings.
First, the team used the three algorithms to predict the 3D structures of the “never born,” shuffled protein sequences from the Czech study. The predictions identified each protein’s structure as folded or disordered in a way that matched the study’s experimental results.
Next, Liu, Yuan and their Israeli mentors applied the algorithms to “newly-born” orphan proteins, very few of which had been purified and adequately characterized experimentally. After searching through the scientific literature, the scientists identified seven such orphan proteins whose function, but not structure, was known.
The AI tools indicated that five of the seven were compactly folded, while two appeared to lack a defined structure. For one of the five, the three algorithms made such strikingly similar predictions—signaling a very high likelihood of accuracy—that the journal featured the three 3D structures on its cover.
In addition, the scientists searched the Protein Data Bank and found three orphan proteins whose crystal structure had been determined experimentally. Remarkably, two of these proteins displayed folds that are not known to exist elsewhere. Since structure determines a protein’s function, the novel folds suggest that some orphan proteins might perform previously unknown biological functions that in the future could be exploited in a host of useful applications, from cutting up plastics to generating clean energy or treating disease.
Says Sussman, “This research changes our idea about how evolution might work. Evolution usually progresses in the way described by Darwin, but occasionally, proteins might appear, in a sense, out of thin air. So, new traits might come out of nowhere, as it were, rather than having evolved from ancestors over millions of years.” Silman adds that the study’s findings, along with other studies on “newly-born” proteins, change the thinking about the origin of life in general, and of humans in particular: “It looks as if we are not just the great grandchildren of E coli.”
Sussman summarizes, “We hope that our study will stimulate other scientists to examine orphan proteins with AI prediction tools to get an idea of their structure and function. When an entirely new structure appears, all bets are off regarding what the protein might be doing biochemically. And that’s when exciting new research horizons open up.”
Liu is now studying for her MSc degree in Prof. Naama Barkai’s lab in Weizmann’s Molecular Genetics Department, and Yuan is currently a graduate student at the University of Texas Southwestern Medical Center, in Dallas. Prof. Sussman is in Weizmann’s Chemical and Structural Biology Department, Prof. Silman, in the Brain Sciences Department and Prof. Binghai Yan, in the Condensed Matter Physics Department. Prof. Amit Finkler, of the Chemical and Biological Physics Department, coordinated the YutChun-Weizmann Program in the Chemistry Faculty.
The YutChun-Weizmann Program is part of an initiative intended to promote academic collaboration between China and the international scientific community. Among its activities, the program provides outstanding undergraduate students with research opportunities.
More information:
Jing Liu et al, Do “Newly Born” orphan proteins resemble “Never Born” proteins? A study using three deep learning algorithms, Proteins: Structure, Function, and Bioinformatics (2023). DOI: 10.1002/prot.26496
Provided by
Weizmann Institute of Science
Citation:
Using three AI protein prediction tools, study uncovers new wrinkles in the folding story of ‘orphan’ proteins (2024, February 14)