Deep learning is a form of artificial intelligence transforming society by teaching computers to process information using artificial neural networks that mimic the human brain. It is now used in facial recognition, self-driving cars and even in the playing of complex games like Go. In general, the success of deep learning has depended on using large datasets of labeled images for training purposes.
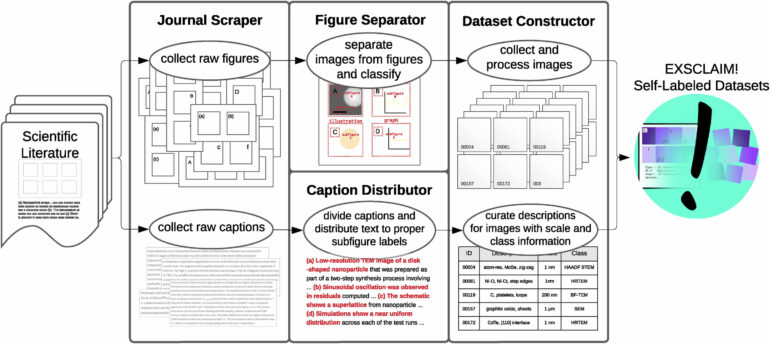
A potential gold mine of labeled images resides within scientific literature, with over a million articles published each year. Most have many figures woven into the text. To date, these figures have not been amenable to deep learning models. This is, in part, due to their complex layouts. Each figure typically contains multiple embedded images, graphs and illustrations. Also lacking has been an adequate means to search the literature for images matching specific content.
Addressing this challenge, researchers at the U.S. Department of Energy’s (DOE) Argonne National Laboratory and Northwestern University have created the EXSCLAIM! software tool. The name stands for extraction, separation and caption-based natural language annotation of images.
The findings are published in the journal Patterns.
“Images generated by electron microscopes down to the billionths of a meter are one of the most important kinds of figures in materials science literature,” said Maria Chan, scientist in Argonne’s Center for Nanoscale Materials, a DOE Office of Science user facility. “These images are essential to the understanding and development of new materials in many different fields. Our goal with EXSCLAIM! is to unlock the untapped potential of these imaging data.”
What sets EXSCLAIM! apart is its unique focus on a query-to-dataset approach, similar to how a prompt is used with generative AI tools such as ChatGPT and DALL-E. It is thus capable of extracting individual images with very specific content from figures, as it both classifies the image content and recognizes the degree of magnification. It can then create descriptive labels for each image. This innovative software tool is expected to become a valuable asset for scientists researching new materials at the nanoscale.
“While existing methods often struggle with the compound layout problem, EXSCLAIM! employs a new approach to overcome this,” said lead author Eric Schwenker, a former Argonne graduate student. “Our software is effective at identifying sharp image boundaries, and it excels in capturing irregular image arrangements.”
EXSCLAIM! has already demonstrated its effectiveness by constructing a self-labeled electron microscopy dataset of over 280,000 nanostructure images. While initially developed around materials microscopy images, EXSCLAIM! is adaptable to any scientific field that produces high volumes of papers with images. The software thus promises to revolutionize the use of published scientific images across various disciplines.
“Researchers now have a powerful image-mining tool to advance their understanding of complex visual information,” Chan said.
More information:
Eric Schwenker et al, EXSCLAIM!: Harnessing materials science literature for self-labeled microscopy datasets, Patterns (2023). DOI: 10.1016/j.patter.2023.100843
Provided by
Argonne National Laboratory
Citation:
New code mines microscopy images in scientific articles (2024, April 9)