CRISPR-Cas systems have revolutionized biotechnology by offering ways to edit genes like a pair of programmable scissors. In nature, bacteria use these systems to fight off deadly viruses. A recent international collaboration led by the University of Copenhagen has shed light on the most enigmatic CRISPR-Cas systems; the type IV system. While these atypical systems do not cut genes, their unique functions show promise in our fight against antibiotic resistance.
CRISPR-Cas systems are bacterial adaptive immune systems that target and cut the nucleic acids (DNA/RNA) of invading genetic parasites like bacteriophages (phages); viruses that infect—and eventually kill—bacterial cells. They consist of two main components; the CRISPR array, which stores immune memory of past viral infections, and the cas genes (encoding Cas proteins), responsible for coordinating the different stages of the immune response.
There are currently six known types of CRISPR-Cas systems, classified according to their protein compositions. All types, except type IV, include nucleases for DNA/RNA cleavage.
CRISPR-Cas systems have gained popularity as gene editing tools, allowing for precise programmable cuts at specific genomic locations—ultimately leading to the 2020 Nobel Prize in Chemistry being awarded for the development of this technology.
Solving the mystery of the missing components
“Type IV systems are the strange cousins among CRISPR-Cas systems, as they lack the immune memory acquisition module and DNA-cutting component that have made CRISPR-Cas systems so famous. These characteristics, and their strict association with mobile circular DNA molecules, called plasmids, motivated us to take on the task of resolving their intriguing role and underlying molecular functions,” explains Fabienne Benz, a University of Copenhagen postdoc and co-first author of a study on this topic published in Cell Host & Microbe.
With the hallmark of CRISPR-Cas being their ability to cut DNA at specific sites, the Type IV systems operate in a totally different way. They lack the typical nuclease “scissors” but harbor a DinG helicase instead—a mysterious protein that unwinds DNA.
“The turning point in this investigation came as we realized that type IV systems do not cut DNA. Instead, we found that they silence gene expression at their target locations. This is a unique functionality that could have important biotechnological applications,” says Rafael Pinilla-Redondo, Assistant Professor at the Department of Biology, and main research coordinator of the investigation.
The researchers reached another breakthrough when they resolved how these systems can function without the necessary components to create immune memory.
“Type IV systems can bypass their lack of a memory acquisition module by hijacking compatible modules from other CRISPR-Cas systems present in the host bacterium. This is fascinating because these other systems are only distantly related,” explains Sarah Camara-Wilpert, co-first author of this study.
Promising CRISPR tool to combat superbugs
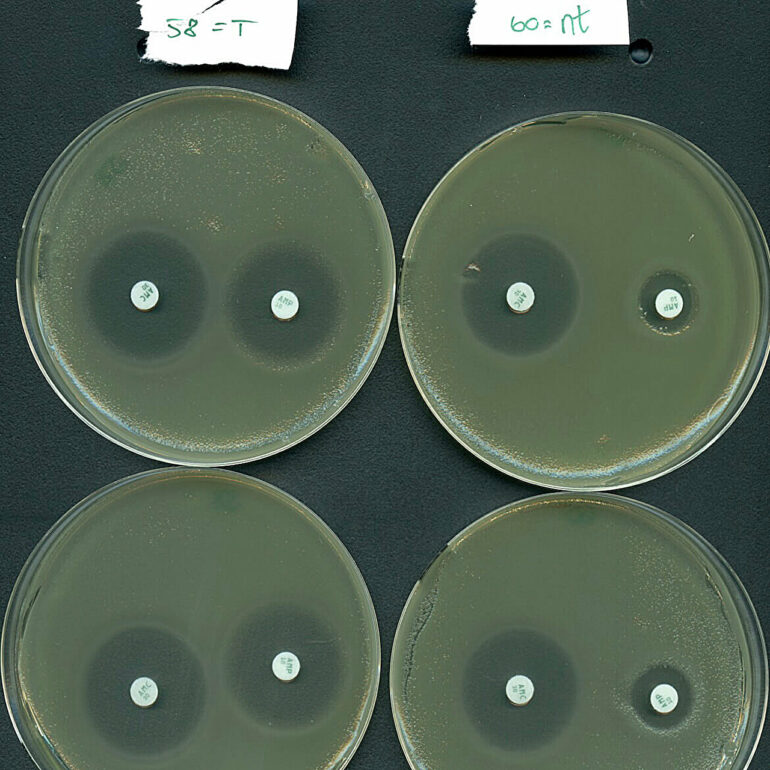
But what’s all the hype about? Well, it turns out that Type IV systems have a marked tendency to naturally target plasmids, rather than bacterial viruses. Importantly, the targeted plasmids frequently harbor multiple antibiotic-resistance genes like those found in hospital superbugs. Antimicrobial resistance is estimated to be directly responsible for over 1 million deaths annually due to treatment failure.
Inspired by their natural plasmid-targeting function, the research teams effectively reprogrammed a type IV system to selectively silence resistance genes carried by a high-risk bacterium from hospitalized patients.
“Our results indicate that the Type IV systems holds potential as a new means to fight antibiotic resistance, as we were able to re-sensitize an important pathogen to antibiotic treatment,” says Professor Søren Sørensen, co-last author of the study.
This study was a major interdisciplinary effort involving seven international research groups from various countries. While the project started as a collaboration between just two groups, it gradually gained momentum, attracting partners with diverse expertise.
“We experienced a wonderful snowball effect, where each new partner amplified the impact of the work by sharing their unique skills and providing crucial insights to solve the mysteries surrounding type IV systems. It has been a collaborative tour-de-force, exactly how science should be,” notes Pinilla-Redondo.
More information:
Fabienne Benz et al, Type IV-A3 CRISPR-Cas systems drive inter-plasmid conflicts by acquiring spacers in trans, Cell Host & Microbe (2024). DOI: 10.1016/j.chom.2024.04.016
Provided by
University of Copenhagen
Citation:
Mystery CRISPR unlocked: A new ally against antibiotic resistance? (2024, May 16)