Superbugs, bacteria that are immune to multiple antibiotics, pose a great challenge to modern medicine. Researchers from the B CUBE—Center for Molecular Bioengineering at TUD Dresden University of Technology and Institut Pasteur in Paris identified a weakness in the bacterial machinery that drives antibiotic resistance adaptation. Their findings, published in the journal Science Advances, could pave the way to boosting the effectiveness of existing antibiotics.
Since the discovery of penicillin in 1928, antibiotics have changed medicine, allowing us to easily combat bacterial infections. However, with the invention of antibiotics, we have also entered a never-ending arms race with bacteria. They adapt rapidly to drugs, rendering many existing treatments ineffective. Such antibiotic-resistant bacteria, often dubbed “superbugs,” pose a critical threat to patients with chronic illnesses and weakened immune systems.
“Rather than developing new antibiotics, we wanted to understand exactly how bacteria adapt their resistances,” says Prof. Michael Schlierf, research group leader at B CUBE, TU Dresden, the leader of the study. In doing so, the groups discovered why it takes longer for some bacteria to develop antibiotic resistance, while others adapt very quickly. Their findings open up new possibilities for the development of counter-strategies.
A genetic toolbox in action
“Our work focuses on the integron system, a genetic toolbox that bacteria use to adapt to their environment by exchanging genes, including those for antibiotic resistance,” says Prof. Didier Mazel, research group leader at Institut Pasteur in Paris, whose group worked together with the Schlierf team.
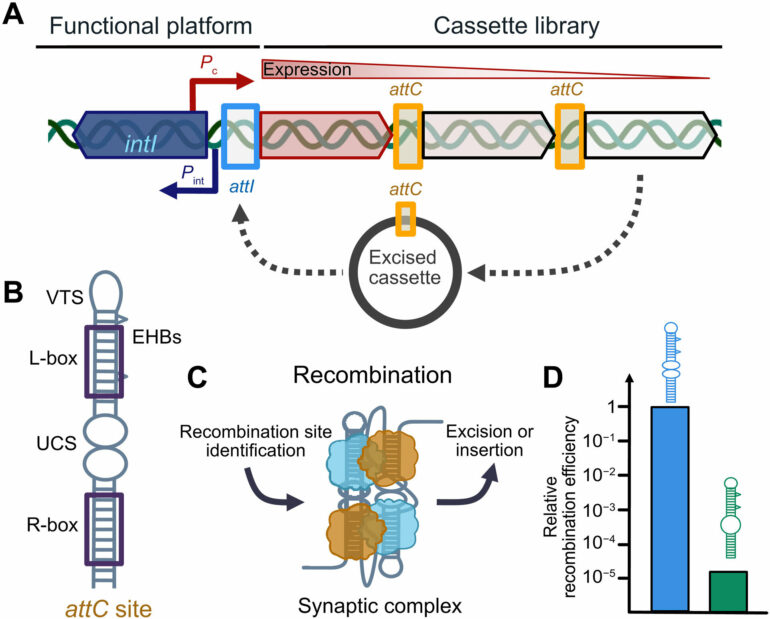
The integron system is like a toolbox. It allows bacteria to store and share resistance genes with their offspring and neighboring cells. It operates via a molecular “cut and paste” mechanism driven by special proteins, known as recombinases. The integron system has been researched a lot. Some bacteria gain new resistance very fast and for others, it takes considerably longer.
It turned out that the variety of DNA sequences is at the heart of this difference. “The sequences inside the integron system are flanked by special DNA hairpins. They are called like this because this is exactly how they look like, like little U-shaped pins sticking out of the DNA. The recombinases are built to bind to these hairpins and form a complex that can then cut out one fragment and paste in another one,” explains Prof. Mazel.
The Schlierf group used a cutting-edge microscopy setup to study how strongly a recombinase protein binds the different DNA hairpin sequences. They found that the complexes with the strongest binding between the protein and the DNA are also the ones that are the most efficient at gaining resistance genes.
Using the force
Using an advanced microscopy technique known as optical tweezers, the Schlierf group measured the tiny forces it takes to pull the different protein-DNA complexes apart.
“With the optical tweezers, we use light to, sort of, grab a single strand of DNA from both sides and pull it apart. Think of it as pulling on a cord to undo a knot,” says Dr. Ekaterina Vorobevskaia, a scientist in the Schlierf lab who carried out the project.
The group saw a clear correlation between the force it took to dismantle a protein-DNA complex and the efficiency of the cut-and-paste machinery.
“If you have a complex that is strongly bound to the DNA, it can perform its job very well. Cut the DNA and paste a new resistance gene very fast. On the other hand, if you have a protein-DNA complex that is rather weak and keeps falling apart, it has to be reassembled again and again. This is why some bacteria gain antibiotic resistance faster than others,” adds Dr. Vorobevskaia.
Exploiting the weakness
“The integron system has been studied by microbiologists for decades. What we bring to the table now is adding the biophysical data and explaining the behavior of this system with physics,” says Prof. Schlierf. “Maybe this vulnerability to force is a more general phenomena for varying efficiencies in biology.”
The scientists believe that the weakness in the system can be used to develop supplemental treatments that will take advantage of, or create, the unstable DNA-protein complexes. It could accompany existing antibiotics and give them an additional time advantage over bacteria.
More information:
Ekaterina Vorobevskaia et al, The recombination efficiency of the bacterial integron depends on the mechanical stability of the synaptic complex, Science Advances (2024). DOI: 10.1126/sciadv.adp8756
Provided by
Dresden University of Technology
Citation:
Scientists find vulnerability in antibiotic resistance mechanism (2024, December 17)