Scientists at the University of California, Irvine, have discovered that nuclear speckles, a membraneless organelle within the nucleus, serve as central hubs for pre-mRNA 3′ end processing. This discovery advances our understanding of how spatial organization within cells impacts gene expression and could open new avenues for treating diseases influenced by RNA processing defects.
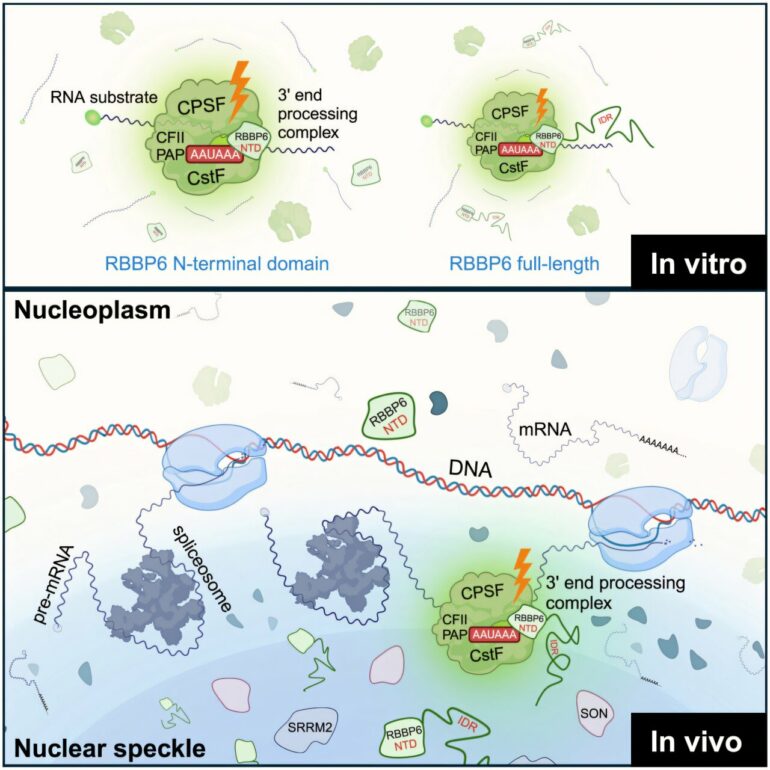
Published in Molecular Cell, the study reveals that RBBP6, a critical factor in pre-mRNA 3′ end processing, localizes to nuclear speckles via its intrinsically disordered region.
This localization is essential for efficient pre-mRNA 3′ end processing in human cells, and the researchers found that more than 50% of genes undergo 3′ end processing at nuclear speckles. These findings highlight the integrative role of nuclear speckles in coordinating transcription, splicing and 3′ end formation.
“Pre-mRNA 3′ end processing is a critical step in mRNA biogenesis, and its misregulation is associated with a variety of diseases, including cancer,” said corresponding author Yongsheng Shi, Ph.D., UC Irvine professor of microbiology and molecular genetics. “Our work, for the first time, identifies where pre-mRNA 3′ end processing occurs in the cell. By understanding this spatial organization, we can better decipher how disruptions in these processes contribute to disease.”
For years, the field has hypothesized the function of nuclear speckles in cells, particularly in the context of other mRNA processing steps, such as splicing. However, testing these functions directly has been challenging due to the complexity of RNA processing machineries.
This new research, led by first author Yoseop Yoon, Ph.D., tackled this challenge by identifying that RBBP6 localizes to nuclear speckles and distinguishing its functional domain from the domain responsible for localization. By making this distinction and employing advanced imaging, sequencing, and biochemical techniques, the researchers were able to directly test whether RBBP6’s localization to nuclear speckles is crucial for pre-mRNA 3′ end processing.
“In addition to its critical role in mRNA processing within nuclear speckles, RBBP6 also binds to many well-known tumor suppressor proteins, such as p53, via its long intrinsically disordered region,” said Shi. “These proteins are known to recruit their target genes to nuclear speckles to enhance gene expression, but the underlying mechanisms remain unknown. Our future work will explore these interactions and their implications in gene regulation and cancer.”
More information:
Yoseop Yoon et al, RBBP6 anchors pre-mRNA 3′ end processing to nuclear speckles for efficient gene expression, Molecular Cell (2025). DOI: 10.1016/j.molcel.2024.12.016
Provided by
University of California, Irvine
Citation:
Nuclear speckles identified as key hubs for gene expression regulation (2025, January 13)