Cells have their own quality control to prevent the production and accumulation of harmful proteins. This quality control is essential for correct embryonic development in all mammals and plays an important role in tumors and genetic diseases such as cystic fibrosis. A group of researchers from the University of Bern and the University of Basel have now made visible and cataloged for the first time, “blueprints” that give rise to defective proteins and are normally recognized and rapidly degraded in cells. This leads to a better understanding of this degradation mechanism and helps in the development of new therapeutic approaches.
When products leave a factory, they must first pass through quality controls. Similarly, when our genes are expressed, there are several such quality controls to ensure that the final products, the manufactured proteins, contain no errors and are functional. In a new study, Professor Oliver Mühlemann and his team at the University of Bern, in collaboration with researchers from the Biozentrum in Basel, have gained new insights into a quality control mechanism that removes defective genetic products from cells, ensuring the error-free expression of our genes. This study has now been published in the journal Genome Biology.
Quality control against defective proteins
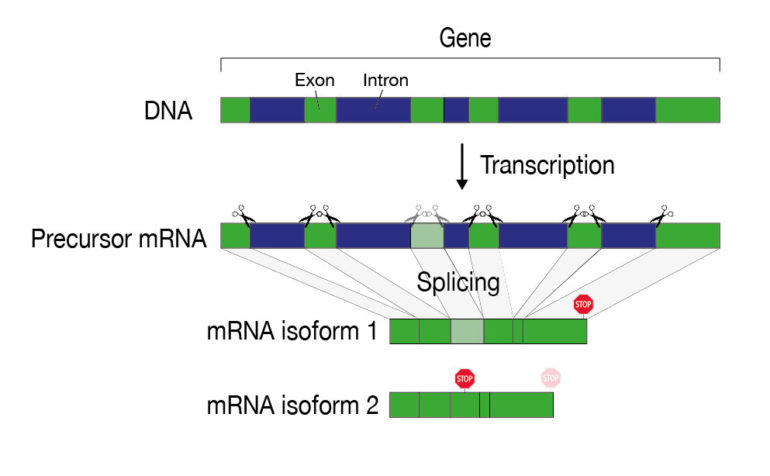
Every cell contains thousands of different proteins, each of which performs a specific task. To produce a specific protein, a gene must first be copied into a molecule of mRNA (messenger RNA). This is then processed and ultimately serves as the blueprint for protein production in the cellular protein factory. “It is important that this mRNA is produced and processed correctly—because if the blueprint contains errors, a faulty protein is produced, which poses a potential risk to the cell,” says Oliver Mühlemann of the Department of Chemistry, Biochemistry and Pharmacy (DCBP) and NCCR RNA & Disease. Therefore, the cell has a number of quality control mechanisms to detect and remove faulty mRNAs. One of these mechanisms, known in technical jargon as nonsense-mediated mRNA decay (NMD), specifically targets mRNAs that contain a code that signals the protein factory to stop the production of a protein too early—even before the protein has been completely produced.
Such defective mRNAs usually emerge during the processing steps that an mRNA must undergo before it serves as a finished template for protein production. One such processing step is splicing, in which certain sequences (the introns) are cut out of the original mRNA and the remaining mRNA (the exons) are spliced back together. This is because in human cells a gene is not present on the DNA as a continuous section, but is interrupted by DNA segments that are not needed for the production of a protein.
The modular structure of a gene allows different mRNA variants and thus also protein variants to arise from one and the same gene. This results in a large number of possible combinations and proteins, which is particularly important for the evolution of complex organisms. However, this process also carries the risk of producing defective proteins.
Degradation products made visible for the first time
In healthy cells, where NMD quality control is active, it is almost impossible to detect defective mRNAs because they are rapidly detected and broken down after production. How then does one determine which mRNAs fall victim to NMD quality control? By eliminating the key players in the quality control process. Thus, the mis-spliced mRNAs accumulate in cells. But here lies the next challenge: these mRNAs come from the same gene as the “correct” mRNA variants and are therefore very similar to them.
With previous sequencing methods, they were almost impossible to distinguish. Previous methods for detecting mRNAs in cells have relied on sequencing many small segments that are later reassembled into a whole using bioinformatics tricks. Evan Karousis of DCBP and NCCR RNA & Disease, first author of the study, explains that the current study used a new method that decodes mRNAs from A to Z in one piece. “This allows each detected mRNA to be uniquely assigned to an mRNA variant,” Karousis said. With these technical tricks, the researchers succeeded for the first time in creating a nearly complete catalog of NMD degradation products in human cells.
A catalog of mRNAs for further research and development of new therapeutic approaches
This classification allows the study of features that distinguish NMD degradation products from “conventional” mRNAs. “If certain mRNAs accumulate that are broken down in healthy cells by NMD quality control, this can contribute to the development of tumors, as is the case in gastric cancer, for example,” Oliver Mühlemann explains. So if we understand how quality control can distinguish defective from correct mRNAs, this knowledge will contribute significantly to developing new therapeutic approaches for diseases in which quality control is impaired.
Gene expression mechanism may have immunity, cancer implications
More information:
Evangelos D. Karousis et al, Nanopore sequencing reveals endogenous NMD-targeted isoforms in human cells, Genome Biology (2021). DOI: 10.1186/s13059-021-02439-3
Provided by
University of Bern
Citation:
Creation of a detailed ‘catalogue’ of degradation products in cells (2021, August 20)
retrieved 23 August 2021
from https://phys.org/news/2021-08-creation-catalogue-degradation-products-cells.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.