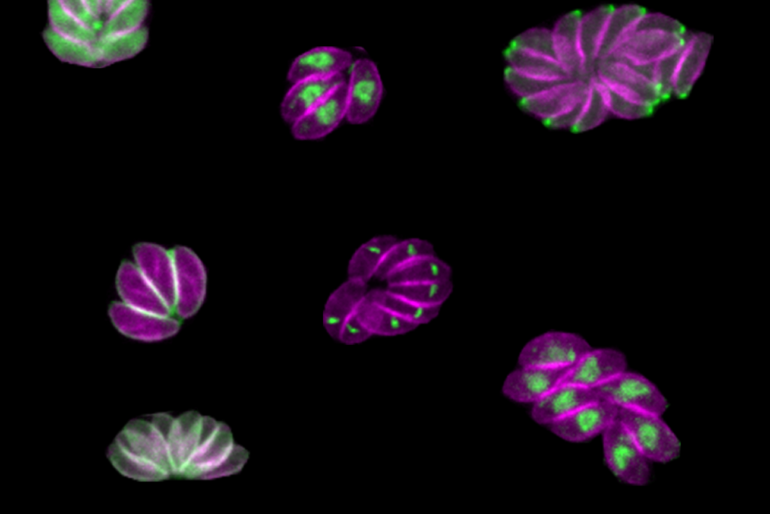
Whitehead Institute Member Sebastian Lourido and his lab members study the parasite Toxoplasma gondii. The parasite causes the disease toxoplasmosis, which can be dangerous for pregnant or immunocompromised patients.
As the parasite evolved over millennia, its phylum (the Apicomplexan parasites) split off from other branches of life, which poses a challenge to researchers hoping to understand its genetics. “Toxoplasma is very highly diverged from the organisms that we typically study, like mice, yeast and [nematodes],” said Lourido lab researcher and Massachusetts Institute of Technology (MIT) graduate student Tyler Smith. “Our lab focuses a lot on developing toolkits to probe and study the genomes of these parasites.”
Now, in a paper published in the journal Nature Microbiology on April 28, Smith and colleagues describe a new method for determining the role of genes within the genome of the parasite. The method can be conducted by a single investigator, and goes a step beyond simply assessing whether or not a given gene is essential for survival. By inserting specific sequences—such as those encoding fluorescent markers or sequences that can turn a gene on and off—throughout the Toxoplasma genome, the method allows the researchers to visualize where an individual gene’s product resides within the parasites and identify when in the life cycle important genes became essential, providing more detailed information than a traditional CRISPR screen.
Although the method could theoretically be used with any gene family, Smith and Lourido decided to first focus on a family of proteins called kinases, the genetic code for which comprises around 150 of Toxoplasma’s 8,000 total genes.
“Kinases are interesting from a basic biology perspective because they are signaling hubs of basic biological processes,” said Smith, who is first author of the study. “From a more translational perspective, kinases are really common drug targets. We have a lot of inhibitors that work with kinases. For some cancers that are linked to specific kinases, the inhibitors can be chemotherapies.”
Using the method, researchers discovered a gene encoding a previously unstudied kinase which they named SPARK. They were able to show that the SPARK kinase is involved in the process of the parasites entering and leaving host cells, and future research on inhibitors of SPARK could lead to new treatments for toxoplasmosis. “Identifying these kinases that are really vital for these critical decision points in a parasite’s life cycle could be really fruitful for developing new therapeutics,” said Lourido, who is also an associate professor of biology at MIT.
New dimensions of screening
Many CRISPR screens use gene editing technology to knock out genes throughout the genomes of a sample of cells, creating a population where every gene in the genome is mutated in at least one of the cells. Then, by looking at which mutations have detrimental effects on the cells, researchers can extrapolate which genes are essential for survival.
But the workings of a whole organism are infinitely more complicated than just survival or death, and researchers are often faced with a challenge when it comes to figuring out exactly what different gene products are doing in the cells. That’s why Smith and Lourido decided to design a method of screening for Toxoplasma genes that could provide more information about what the products of those genes do. “CRISPR screens can tell you which genes are important, but it doesn’t give you much information about why they’re important,” Smith said. “We were seeking to make a kind of platform that could look at other dimensions.”
Smith and Lourido used CRISPR technology to introduce small amounts of new DNA into the parasites’ genes that code for kinases. The new DNA included sequences encoding a fluorescent marker protein and sequences that could be used to manipulate gene expression levels.
After creating a population of parasites modified this way, the researchers then used imaging to determine where the fluorescently tagged proteins had ended up in the cells, and to observe what happened in the cells when the proteins were turned off. “Being able to see different cell division phenotypes—for instance parasites that either failed to replicate at all, or tried to replicate but would have some abnormalities—that gets us closer and allows us to generate hypotheses as to actually why these kinases are important, not just whether or not they are important,” Smith said.
The depletion of some proteins caused the parasites to die instantly, while others affected the parasites at a later point in their life cycles, so they would drop out of the population more slowly. “Cells with mutations in these kinases replicate fine, but a problem might arise when they need to leave their host cell and enter a new host cell later on down the line,” Smith said.
A ‘SPARK’ of inspiration
After the screen, the researchers followed up on one of these kinases in particular, which they called SPARK (short for Store Potentiating/Activating Regulatory Kinase). Mutants depleted of SPARK died, but not until a later phase of the life cycle. Smith and Lourido conducted further experiments to understand SPARK’s role, and found evidence that the protein was involved in the release of calcium in the cell that is required for a parasite to enter or leave a host cell.
“The thing I found very interesting about SPARK is that it’s a kinase that’s very different from the analogous kinase in other model organisms, but is conserved throughout all of the apicomplexan phylum,” Smith said. “That’s the phylum that includes Toxoplasma and a bunch of other single-celled parasites like Plasmodium, which is the malaria parasite.”
Because SPARK is far different from its human analog and essential to the parasite’s life cycle, a SPARK-specific kinase inhibitor could be used to treat toxoplasmosis by killing the parasite without affecting the patient. “The hope would be that you can target SPARK and inhibit it without hitting mammalian kinases,” Smith said. “It’s easy enough to design something that kills a cell, but the trick is only killing parasites and not your own cells.”
In the future, the researchers hope to turn their new screening method to other families of genes, such as transcription factors, to understand their function in the parasites. “Our results have been quite encouraging in that we think this method will be scalable, and we can target larger gene sets in the future,” Smith said. “I think the ultimate end goal would be to do the whole genome.”
“There’s this whole universe of parasite proteins that we know so little about, where this type of analysis will be incredibly insightful.” Lourido said. “We’re really very excited about scaling it up further.
More information:
Tyler A. Smith et al, Screening the Toxoplasma kinome with high-throughput tagging identifies a regulator of invasion and egress, Nature Microbiology (2022). DOI: 10.1038/s41564-022-01104-0
Provided by
Whitehead Institute for Biomedical Research
Citation:
Novel screening approach reveals protein that helps parasites enter and leave their hosts (2022, April 29)