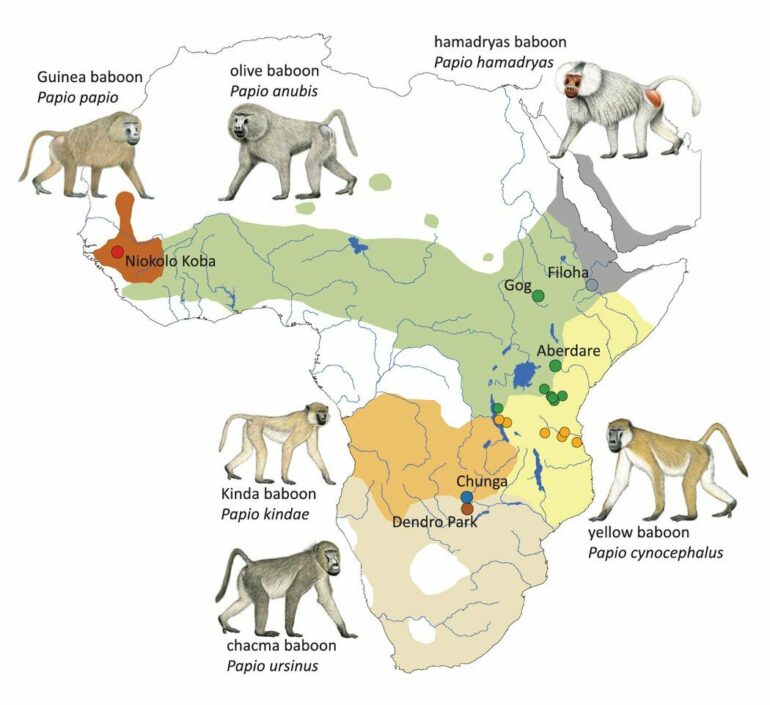
Baboons (Papio) are found across the continent of Africa, from the west to the east and all the way south. They have doglike noses, impressive teeth and thick fur that ranges widely in color between the six species, which are olive, yellow, chacma, Kinda, Guinea and hamadryas. Their habitats vary from savannas and bushlands to tropical forests and mountains.
Chacma baboons, the largest at up to 100 pounds, are even found in the Kalahari Desert, while the neighboring Kinda baboons, the smallest at around 30 pounds, stay near water. Most live in large troops with dozens or hundreds of members. While most baboons are polygynandrous, with males and females mating with multiple partners, hamadryas baboons, also called sacred baboons, live exclusively in units of one male and multiple females.
In a paper published today in the journal Science, titled “Genome-wide Coancestry Reveals Details of Ancient and Recent Male-driven Reticulation in Baboons,” researchers show surprising amounts of genetic admixture between baboon species, something that also likely occurred in early humans. Mark Batzer, Boyd Professor and the Dr. Mary Lou Applewhite Distinguished Professor of Biological Sciences at LSU; Jessica Storer, Ph.D., Batzer’s former student at LSU and now research scientist; and LSU research associate Jerilyn Walker all contributed to the research. Together, they analyzed the mobile or “transposable” genetic elements in samples from 225 baboon individuals from 19 geographical sites.
“Everybody believes their genome is perfectly stable, and that’s exactly wrong,” Batzer said. “Well over half of the genome is fluid in nature and moves around in and between individuals, and between generations and populations. This mobile part of the genome, or mobilome, provides important clues as to how different species are related to one another, how they differ and when two individuals share a common ancestor.”
Whole-genomic sequencing has revolutionized the amount and detail of genetic diversity now available to researchers to study. While the LSU researchers previously had looked at a few hundred mobile elements or “jumping genes,” primarily of the Alu and L1 types, they were now able to analyze over 200,000 elements computationally, confirming and expanding on previous studies. The broader research consortium includes more than 30 collaborators around the globe and was led by Jeffrey Rogers, associate professor of molecular and human genetics at Baylor College of Medicine.
“There are questions that were science fiction when I started in the field that are now perfectly approachable,” Batzer said. “We’re also brought back to this fundamental question, ‘What even is a species?’ When I was a young scientist, it meant reproductive isolation; no exchange of genes back and forth, and individuals from different species would form infertile hybrids. Well, that whole concept has evolved, and what we now see are free exchanges of genes back and forth, both in ancient times and more recently. In other words, there hasn’t been a linear trajectory of genetically isolated species that change through time.”
Mobile, transposable elements cause a subset of all genetic mutations known as structural genetic variants, one of the most important types of mutation in the genome. As such, mobile elements are responsible for some genetic diversity, but not all differences. Their activity, or rate of movement, is also variable between species, including at different times. While baboons currently are on “fast forward,” orangutans, for example, are almost on pause. Humans are somewhere in between.
“You can say mobile elements like Alu and L1 are involved in a genetic arms race or competition within the genome,” Batzer said. “The mobile elements attempt to expand in number, while the genome exerts control over that expansion, so the elements don’t ‘overrun’ the genome and cause so much havoc it risks killing the host. Some mobile elements are distant relatives of viruses, so some of the control systems are the same ones that control the spread of viruses.”
Apparent similarities, such as between two individuals of the same species, can disguise surprising amounts of genetic diversity, as one baboon can have almost as much in common—genetically speaking—with a baboon from a different species. The researchers were also able to show, for the very first time in non-human primates, how the yellow baboons in western Tanzania received genetic input from three distinct lineages—yellow, olive and Kinda.
“This was the first time we’ve seen three different species contribute to the genesis of one, and done it in detail,” Batzer said. “These high-resolution data sets allow us to draw much more accurate and detailed conclusions from the observations we make.”
Baboons and humans share about 91 percent of identical DNA. While humans have relatively small amounts of variation from each other, baboons are genetically more diverse. Bigger mobile elements called LINE elements, such as L1, carry around enzymatic machinery that helps them and the smaller Alu elements mobilize and drive change in mammals (L1) and primates (Alu).
Mobile and transposable elements are in themselves diverse and effectively “monkey around” the genomes of all primates, including humans, as well as other species. The processes by which they impact the genome are called insertional mutagenesis, transduction and recombination. Tracking the insertions, which is Batzer’s specialty, offers two advantages in establishing shared or separate ancestry.
First, the presence of a mobile element at a particular location in the genome represents identity by descent; the probability of an exact match without shared ancestry is near negligible. Second, it’s possible to trace insertions back to the point where they first appeared, thus establishing the ancestral genetic character state and unambiguously rooting species relationships.
“We now believe mobile elements are one of the single biggest driving forces impacting genomes, and not just among primates, but across many mammals and many non-mammalian systems as well,” Batzer said.
Next, the LSU research team will investigate the mobilization and genomic impact of a recently identified transposable element in South American primates.
More information:
Erik F. Sørensen et al, Genome-wide coancestry reveals details of ancient and recent male-driven reticulation in baboons, Science (2023). DOI: 10.1126/science.abn8153
Provided by
Louisiana State University
Citation:
Researchers show mobile elements monkeying around the genome (2023, June 1)