There are many types of cancer treatments. But which ones work best varies from patient to patient. Currently, doctors determine which treatment to try for a patient based on where in their DNA, or genetic code, the error that caused the cancer is located.
But a new approach that groups patients by the changes in protein structure and function caused by that error, rather than by the location of the changes in DNA, could lead to both more inclusive clinical trials and better treatment matches for patients.
I am part of a team that researches targeted therapies for cancer and ways to make treatment more patient-specific. A recent study by our research team determined that grouping DNA errors by structure may better personalize cancer therapies.
Targeted therapies hone in on cancer cells
Mutations are errors made during cell replication when the genetic material of that cell, or DNA, makes a new copy of itself. These mutations are usually harmless and caught by the cell’s proofreading machinery.
However, the proofreader occasionally fails. And on rare instances, these mutations occur in parts of the DNA called oncogenes. Under normal conditions, oncogenes are essential to normal growth and development, such as fetal organ development and general tissue repair and maintenance. But when mutations cause oncogenes to signal for unregulated growth, cancer can form.
One way to kill these tumor cells is to use targeted therapy. Cancer-targeted therapies bind specifically to the defective proteins produced by mutated oncogenes and prevent them from sending a “grow” signal.
Because targeted therapies bind directly to the cancerous protein, they spare most noncancerous cells from being harmed. This results in more specific cancer cell killing and less overall treatment toxicity. In contrast, chemotherapy attacks all actively dividing cells, which includes not just cancer but also hair follicles, the digestive tract and other parts of the body.
To make targeted therapies more specific, scientists often study the physical or structural changes that mutations cause in proteins. They design drugs that preferentially bind to these particular changes to prevent the defective protein from causing uncontrolled growth.
However, because mutations can occur in many different areas of a protein, multiple targeted therapies are often necessary to bind all the different mutations that occur across cancer types. This leads to a difficult clinical problem: How do physicians match patients to the most effective targeted therapy for their mutations?
Traditional treatments use mutation location
To attempt to answer this question, our research team chose to focus on one oncogene in lung cancer, EGFR, or epidermal growth factor receptor. We did this for two reasons. First, lung cancer remains the No. 1 cause of cancer-related deaths worldwide. Second, EGFR mutations are among the most common forms of lung cancer – they occur in approximately a third of non-small-cell lung cancers worldwide, amounting to over 550,000 patients annually.
F. Gervasio/UCL Chemistry and ISMB, CC BY
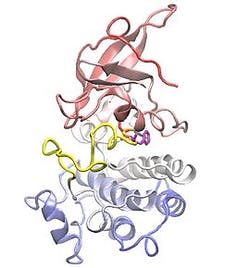
EGFR has many different mutations that cause unregulated growth. Multiple generations of targeted therapies are available for this population of patients.
Clinical trials and treatment options for patients with oncogene-driven lung cancer, particularly with EGFR, are currently based on the type and location of the mutation in the DNA.
However, mutation location is not the best approach to predict how well patients will respond to a drug. Because mutations change the shape of a protein, they can alter the way targeted therapies interact with the protein.
Regrouping cancer mutations
By examining the mutated structures of different EGFR proteins, our team found that they could be categorized into distinct subgroups.
For example, we found that mutations that form away from the areas of the protein that drugs target don’t noticeably affect how well the drug binds to the protein. Cells with this kind of protein mutation were thus killed by all types of EGFR inhibitors. Although these mutations occurred across many locations in the DNA, they shared the same overall structural and functional effect on proteins.
Conversely, mutations that form near typical drug target areas compress this region and prevent certain EGFR inhibitors from attaching to the protein. These mutations also occurred in several different DNA locations.
rightdx/iStrock via Getty Images Plus
Based on these findings, our team hypothesized that structural changes in similar protein regions, not DNA location, would cause similar changes in how well a drug works.
To test our hypothesis, we retrospectively analyzed public and hospital data on how well patients responded to cancer treatments. We sorted patients into traditional DNA location-based groups and our newly defined structure/function-based subgroups to determine whether one group had more patients who responded better than the other to different drugs.
We found that the structure/function-based subgroups identified nearly twice as many patients that benefited from a particular drug compared with the DNA location-based groups. Grouping patients by structure/function also identified which EGFR inhibitor provided the longest clinical benefit for patients.
More inclusive clinical trials
In addition to potentially matching patients to more effective treatments, clinical trials using structure-based subgroups may lead to broader access to therapies.
Current clinical trials exclude up to a fifth of patients with EGFR mutant non-small-cell lung cancer because each clinical trial typically focuses on only a handful of specific mutation types. Reframing clinical studies to be based on the changes that mutations cause to protein structure and function, as opposed to their location on DNA, could expand treatment options to include patients with more rare EGFR mutant cancers.
This approach provides a framework that clinical trials could use to make studies more inclusive of all types of mutations. And it may also identify previously ignored or hidden mutation subgroups that can lead to additional drug development and ultimately improve patient care.
[Understand new developments in science, health and technology, each week. Subscribe to The Conversation’s science newsletter.]