Predicting a protein’s location within a cell can help researchers unlock a plethora of biological information that’s critical for developing future scientific discoveries related to drug development and treating diseases like epilepsy. That’s because proteins are the body’s “workhorses,” largely responsible for most cellular functions.
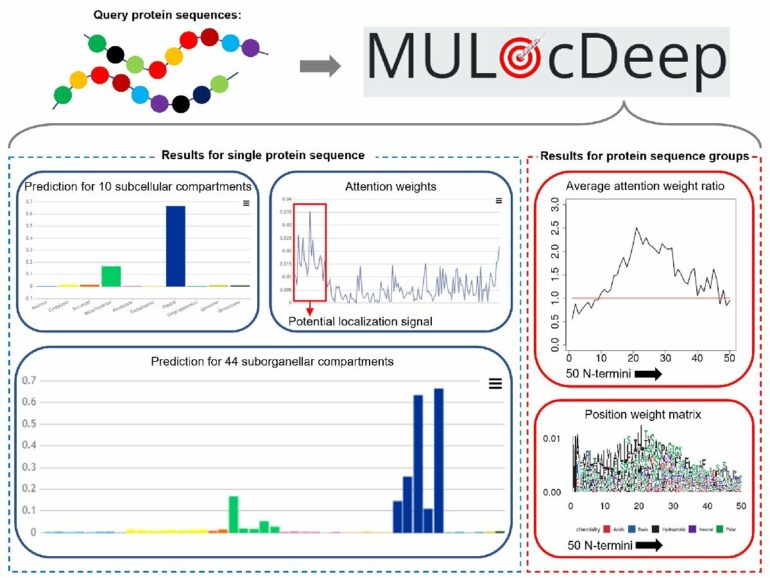
Recently, Dong Xu, Curators Distinguished Professor in the Department of Electrical Engineering and Computer Science at the University of Missouri, and colleagues updated their protein localization prediction model, MULocDeep, with the ability to provide more targeted predictions, including specific models for animals, humans and plants. The model was created 10 years ago by Xu and fellow MU researcher Jay Thelen, a professor of biochemistry, to originally study proteins in mitochondria.
“Many biological discoveries need to be validated by experiments, but we don’t want researchers to have to spend time and money conducting thousands of experiments to get there,” Xu said. “A more targeted approach saves time. Our tool provides a useful resource for researchers by helping them get to their discoveries faster because we can help them design more targeted experiments from which to advance their research more effectively.”
By harnessing the power of artificial intelligence through a machine learning technique—training computers to make predictions using existing data—the model can help researchers who are studying the mechanisms associated with irregular locations of proteins, known as “mislocalization,” or where a protein goes to a different place than it’s supposed to. This abnormality is often associated with diseases such as metabolic disorders, cancers and neurological disorders.
“Some diseases are caused by mislocalization, which causes the protein to be unable to perform a function as expected because it either cannot go to a target or goes there inefficiently,” Xu said.
Another application of the team’s predictive model is assisting with drug design by targeting an improperly located protein and moving it to the correct location, Xu said.
In the future, Xu hopes to increase the model’s accuracy and develop more functionalities.
“We want to continue improving the model to determine whether a mutation in a protein could cause mislocalization, whether proteins are distributed in more than one cellular compartment, or how signal peptides can help predict localization more precisely,” Xu said. “While we don’t offer any solutions for drug development or treatments for various diseases per se, our tool may help others for their development of medical solutions. Today’s science is like a big enterprise. Different people play different roles, and by working together we can achieve a lot of good for all.”
Xu is currently working with colleagues to develop a free, online course for high school and college students based on the biological and bioinformatics concepts used in the model and expects the course will be available later this year.
A conflict of interest is also noted by Xu and colleagues: While the online version of MULocDeep is available for use by academic users, a standalone version is also available commercially through a licensing fee.
“MULocDeep web service for protein localization prediction and visualization at subcellular and suborganellar levels,” was published in the journal Nucleic Acids Research. Co-authors are Yuexu Jiang, Lei Jiang, Chopparapu Sai Akhil, Duolin Wang, Ziyang Zhang and Weinan Zhang at MU.
More information:
Yuexu Jiang et al, MULocDeep web service for protein localization prediction and visualization at subcellular and suborganellar levels, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad374
Provided by
University of Missouri
Citation:
AI software can provide ‘roadmap’ for biological discoveries (2023, June 2)