Cells can now be genetically programmed to record their histories within their genomes, a development that could revolutionize the study of developmental and disease processes, according to a collaborative work by researchers from multiple institutions including the University of California, Los Angeles and University of Washington, Seattle.
Non-invasive monitoring of basic physiological variables through methods such as urine or blood sampling offers a signal of processes occurring somewhere in the body. Additional testing of tissue samples and clinical assessments are then used to diagnose or treat based on the evidence that further testing can offer.
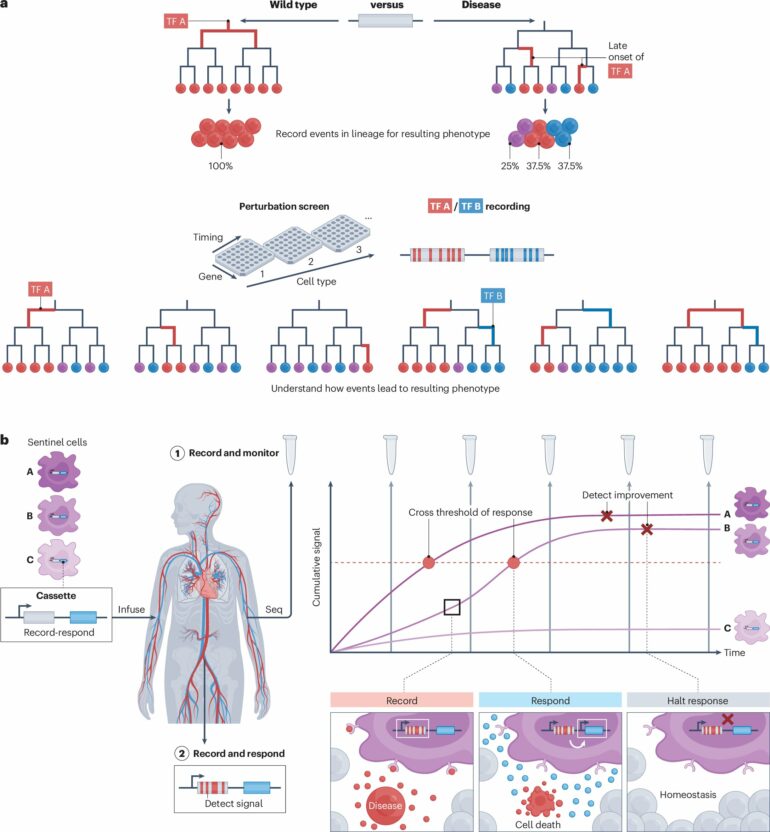
A fundamental challenge in research is explaining biological states or processes in terms of preceding events. Cellular histories encompass lineage relationships, extrinsic influences, internal states, and spatial contexts over time.
Methods for obtaining these histories face reliability limitations due to selection bias, scalability and the destructive nature of some measurement techniques, essentially allowing researchers to see only snapshots of the current state of a cell without a history of how it got there. Traditionally, this requires multiple snapshots over time to construct an evolution of events, with each snapshot facing similar selection bias limitations.
Techniques such as site-specific recombinases, CRISPR systems, and other genome-editing technologies allow cells to write information into their genomes. Recent advances in single-cell and spatial omics technologies enable the recovery of this recorded information.
Having access to recorded molecular and physiological states over time in individuals would provide rich datasets that enable early diagnosis, improved treatment of disease and better-informed health decisions, as well as insight into mechanisms and causes of disease.
The Perspective, “The lives of cells, recorded,” published in Nature Reviews Genetics, discusses how recent advances in genome engineering enable DNA-based recording with the potential to create engineered “memory sentinel cells.”
Sentinel cells could be engineered to capture a diverse range of molecular and cellular biomarkers and then express the information as encoded proteins or nucleic acids which could be