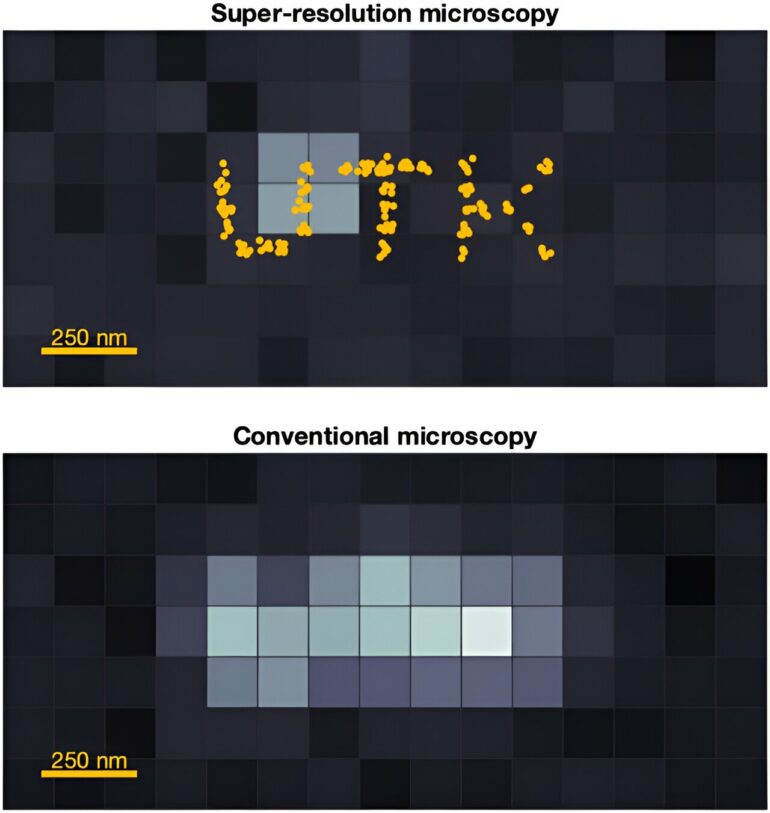
It’s about to get easier to catch and analyze a high-quality image of fast-moving molecules. Assistant Professor Ioannis Sgouralis, Department of Mathematics, and colleagues have developed an algorithm that adds a new level to microscopy: super-resolution in motion.
The cutting-edge advancement of super-resolution microscopy was recognized with the 2014 Nobel Prize in Chemistry for its groundbreaking innovation. It improves optical microscopy with a suite of techniques that overcome the inherent limitations set by the physics of light. The high-frequency oscillations of light waves escape detection by the naked eye or conventional cameras, appearing continuous. Super-resolution microscopy captures details more refined than the wavelength of light which, due to diffraction, are otherwise missed by common microscopes and optical devices.
“For scientific experiments in biochemistry and molecular biology, where we typically need to observe individual biomolecules, such missing details are critical,” said Sgouralis. “Characteristically, important biomolecules like DNA, RNA, and proteins are about 1,000 times smaller than light’s wavelength, as a result their images appear noisy, distorted, and heavily blurred—which makes them inappropriate for scientific purposes.”
Super-resolution tools such as PALM or STORM fill in these details by relying on image-analysis algorithms to recover the missing information and capture accurate still images at the molecular level.
“Although super-resolution experiments have had a huge impact on the life sciences, they allow recovery of the missing information only when the biomolecules remain immobile,” said Sgouralis. “However, life is all about motion and biomolecules within a living organism are constantly moving.”
In their new research, published July 22 in Nature Methods, Sgouralis and colleagues demonstrate a new framework called Bayesian nonparametric track (BNP-Track), the first image-analysis algorithm that allows super-resolution for moving biomolecules.
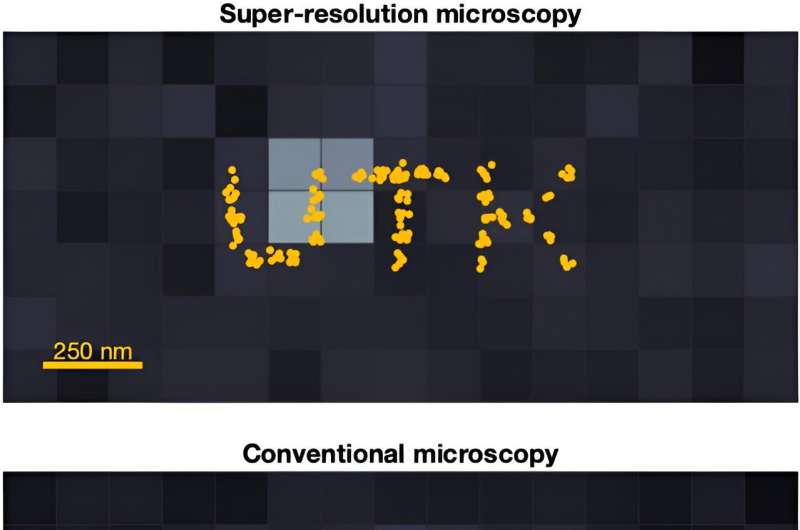
Super-resolution microscopy reveals intricacies unseen by conventional imaging, bringing minute details into exquisite focus, opening up the molecular nano-cosmos. © University of Tennessee at Knoxville
“We developed advanced mathematical methods that analyze images of a microscopy experiment and recover the missing information even when the biomolecules are constantly changing position,” said Sgouralis. “Our work allows direct observation of moving biomolecules within living cells and the reconstruction of their motion with much finer accuracy than is provided within the wavelength of light. This now enables innovations in biochemistry, molecular biology, and biotechnology that were previously inaccessible.”
BNP-Track allows researchers to address unanswered questions about biomolecular behavior: Do biomolecules tend to aggregate in certain locations within a cell? Do they originate from one or multiple locations? How are they transported from a cell’s exterior to the cell’s interior or from cell to cell? Do certain biomolecules prefer to stay together or fall apart?
“These are only some of the questions that are typically asked in drug discovery or when studying the central dogma of molecular biology,” said Sgouralis.
Next steps for BNP-Track research will seek to condense the time needed to run these new algorithms.
“To analyze images of one experiment only, they need to run for several hours,” said Sgouralis. “Research in the immediate future needs to reduce these to maybe a few minutes or seconds. Then, we can analyze images from multiple experiments quickly.”
They will also develop specialized versions of the algorithms to work with the variety of microscopy setups needed across the spectrum of laboratory situations. In the meantime, the BNP-Track innovation by Sgouralis and team provides a new foundation for discovery.
More information:
Ioannis Sgouralis et al, BNP-Track: a framework for superresolved tracking, Nature Methods (2024). DOI: 10.1038/s41592-024-02349-9
Provided by
University of Tennessee at Knoxville
Citation:
BNP-Track algorithm offers a clearer picture of biomolecules in motion (2024, August 2)