Gene regulation underpins nearly every biological process—from cell development to responses to environmental changes, and understanding it can provide insights into cancer and other diseases. Now, FMI researchers have made significant progress in uncovering how transcription factors—proteins that control gene expression—work in fission yeast, a key model organism for studying gene regulation.
The team’s research is published in the journal Molecular Cell.
One of the main players in gene regulation are transcription factors—proteins that control the expression of genes by binding to specific regions of DNA. These proteins act as switches, regulating the level of expression of genes in response to various signals.
Scientists have learned a lot about how genes are controlled by studying simple organisms such as fission yeast (Schizosaccharomyces pombe). However, most transcription factors haven’t been fully studied, and there’s still much to learn about how these proteins work with DNA and other proteins to regulate gene activity.
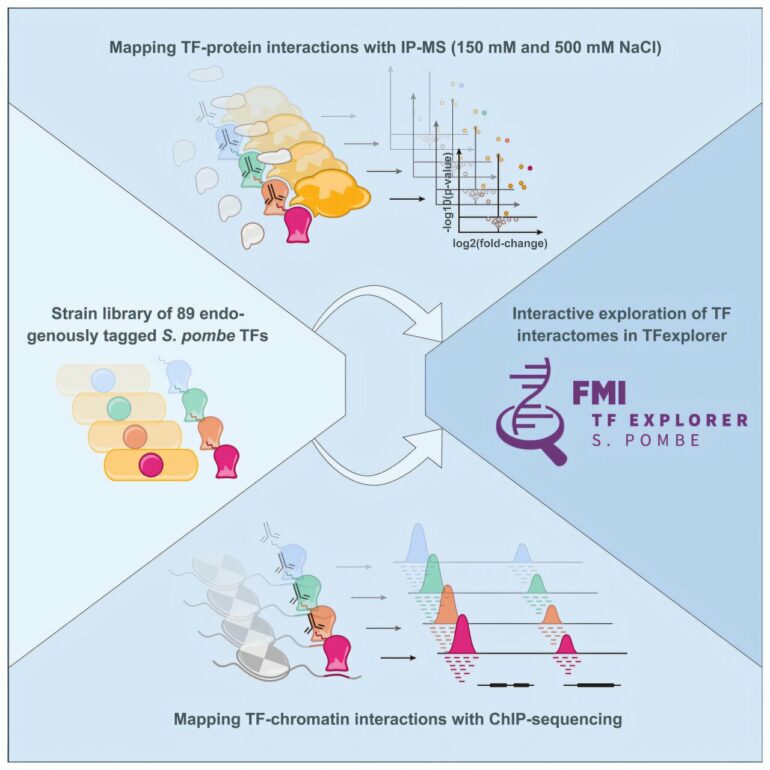
To explore how transcription factors control gene expression, researchers led by Merle Skribbe, a Ph.D. student in the Bühler lab, have used genome editing to create 89 different strains of fission yeast. In each strain, they tagged a specific transcription factor with a marker that made it easier to study these proteins individually and map their interactions with other proteins and DNA.
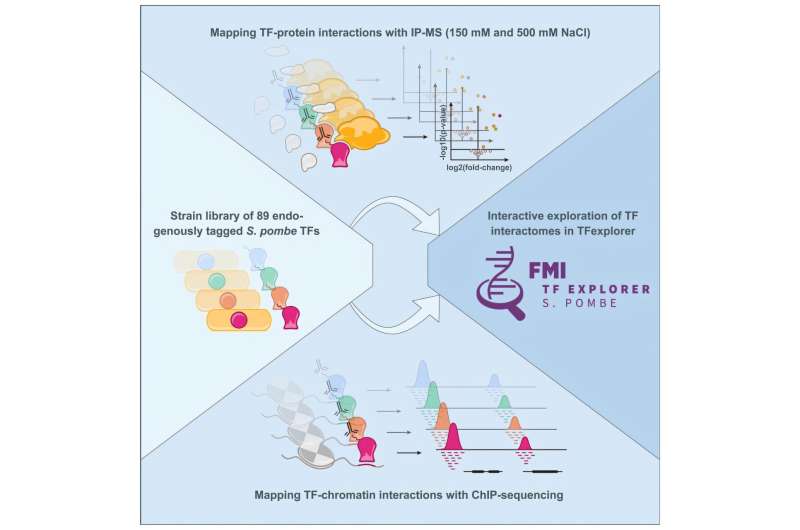
Graphical abstract. © Molecular Cell (2025). DOI: 10.1016/j.molcel.2025.01.032
The researchers identified “HOT” regions—sections of the DNA that are bound by many different transcription factors—and discovered new interactions. These included interactions with certain regulatory proteins, suggesting there might be a shared method across organisms for controlling transcription factor activity. The findings advance our understanding of how genes are regulated in fission yeast but could also reveal insights into gene regulation in other organisms, Skribbe says.
A major finding was the discovery of a pair of transcription factors that the researchers dubbed the “Nattou complex.” This complex appears to regulate gene expression and influence the positioning of certain genes within the nucleus—a finding that could provide a better understanding of gene silencing mechanisms, which are essential for regulating development, immune responses and other important biological processes.
The work also led to the creation of FMI TFexplorer, a free online tool that lets scientists explore transcription factor interactions in an easy and interactive way—no advanced computer skills required.
Bühler emphasizes the importance of teamwork between his group and the technology platforms at the FMI. Researchers from the Functional Genomics and the Proteomics and Protein Analysis platforms helped to collect the data, and Skribbe worked with Charlotte Soneson and Michael Stadler from the Computational Biology platform to analyze it.
Bühler also praised the leadership of Skribbe, noting her extraordinary independence and leadership. “For this, I offered her co-corresponding authorship on the paper—a rare distinction at her career stage,” he says.
The study represents a significant step forward in understanding the role of transcription factors, Bühler adds, with the open-access data offering a powerful resource for scientists exploring the complexities of gene regulation.
More information:
Merle Skribbe et al, A comprehensive Schizosaccharomyces pombe atlas of physical transcription factor interactions with proteins and chromatin, Molecular Cell (2025). DOI: 10.1016/j.molcel.2025.01.032
Provided by
Friedrich Miescher Institute for Biomedical Research
Citation:
Charting gene secrets: Fission yeast strains provide insights into genetic switches (2025, February 26)