Researchers in the lab of John Kuriyan, professor of biochemistry and dean of the School of Medicine Basic Sciences, have revealed a key element of how a molecular machine responsible for high-speed DNA replication works. The results of their study build on growing theories of molecular evolution.
The article, “Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM,” was published in the journal Nature Structural & Molecular Biology.
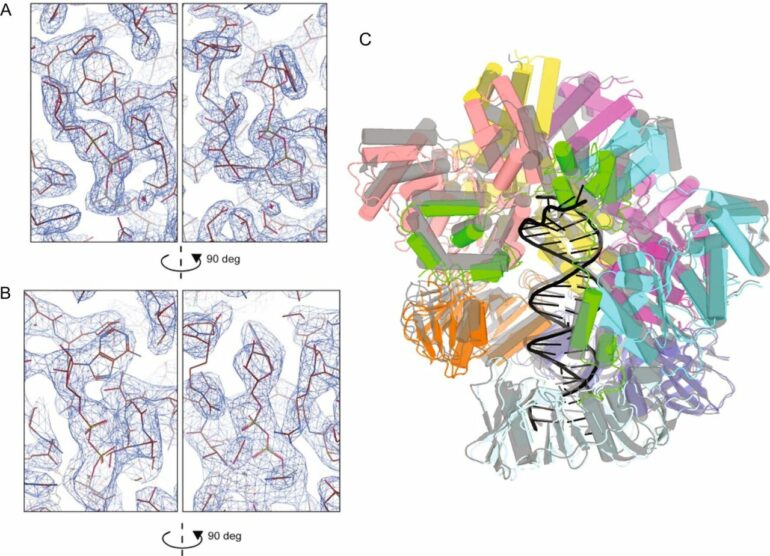
Clamp loaders—proteins that help load DNA replication machinery onto the DNA—have a variety of biological functions that change as their structure evolves. These proteins are found in all of life—from single-celled organisms to humans. Over the last decade, the structure and function of the clamp loader were thought to be well understood based on a handful of snapshots of its three-dimensional structure. This research, led by co-first authors postdoctoral fellow Kendra Marcus and research assistant professor Yongjian Huang, revealed the bigger picture.
“By combining deep mutagenesis and cryogenic electron microscopy , we found a key structural intermediate that was not seen in these [snapshots] and illustrated the conformational changes needed for the clamp loader to do its job,” Marcus said. “Not only this, but we see regions of the protein that we thought were ‘unimportant’ participate in highly coordinated processes.”
The unique pairing of deep mutagenesis and cryo-EM is what led to this discovery. Deep mutagenesis maps important regions of a protein by revealing its mutational sensitivity, or how a protein responds to a mutation, and cryo-EM produces three-dimensional visualizations that aren’t usually seen in crystal structures. Studying the mutational sensitivity of proteins sheds light on how the protein architecture influences its function and how evolution can tune these variables to give proteins new tasks.
The researchers first inferred a new state of the clamp loader based on the deep mutagenesis data and then visualized it using cryo-EM, which revealed a previously unseen conformational state, or arrangement of atoms that gives the clamp loader its shape.
“One of the unique strengths of cryo-EM lies in its power to capture the ensemble of conformational states adopted by these macromolecular machines, such as the clamp-loader,” Huang said. “In this study, cryo-EM allows us to visualize the molecular details of a newly discovered conformational state and the critical conformational changes required for the function of clamp loader.”
These findings have implications in evolutionary theory and how proteins use ‘insignificant’ places in these molecular machines to become conditionally important in the adaptation of new functions, Marcus said.
Following this discovery, the Kuriyan lab will leverage machine learning techniques to design and test new clamp loader proteins based on their current findings. These types of experiments offer tremendous utility toward understanding how proteins change function because of disease-related mutations or in response to drugs.
“We hope that these results inspire biochemists and molecular biophysicists to apply creative methods to explore the physical space of proteins,” Marcus said. “This work illustrates how evolutionary theory can elevate our structurally oriented understanding of protein functions.”
More information:
Kendra Marcus et al, Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM, Nature Structural & Molecular Biology (2024). DOI: 10.1038/s41594-023-01177-3
Provided by
Vanderbilt University
Citation:
Combining unique methods, researchers discover new protein functions, explore physical space of proteins (2024, February 9)