A model from physics can provide insight into differences between slow and fast muscles, according to new research.
In physics, geometric frustration is a phenomenon that occurs when the minimum-energy (or most desirable) state of a physical system can be achieved by many different arrangements. For example, in the packing of water molecules in ice, there are many different arrangements of hydrogen bonds between the molecules that achieve a minimum-energy configuration.
This can be compared to trying to arrange guests around a dining table in the most favorable seating plan, taking into account friendships and those who don’t get along. If there is more than one arrangement that minimizes the number of disagreeable pairings, the dining table is said to be “geometrically frustrated” (or “frustrated” for short). The guests are frustrated in that if they try to move from one of these optimal arrangements they end up, at best, in an arrangement that is no better.
Frustration is an important concept in physics and has led to a range of developments such as new nanomaterials and electronic devices. Now, an international team including academics from Imperial and the University of Canterbury in New Zealand has shown for the first time that frustration is also present in a native biological system: vertebrate muscle.
Dr. Pradeep Luther, from Imperial’s Faculty of Medicine said: “This is a unique discovery at the interface of physics and biology, helping to explain some old and intriguing observations from electron micrographs of muscle transverse sections. The results appear to be related to the different mechanical properties of different kinds of muscle fiber.”
The study, published in the Journal of The Royal Society Interface, shows that the pattern of protein filament orientations in many types of vertebrate muscle fits well to a fundamental frustrated system in statistical physics.
Lead author Professor Rick Millane said: “This is the first time that geometric frustration has been observed in a native biological system and could provide crucial insight into the difference between slow and fast fatigable muscles.”
About the findings
Muscle is made up of thousands of parallel bundles of muscle fibers, and within each fiber there are thousands of intricately ordered arrays of the contractile proteins actin and myosin. It is the interactions and movements between filaments of these two proteins that is the molecular basis of muscle contraction.
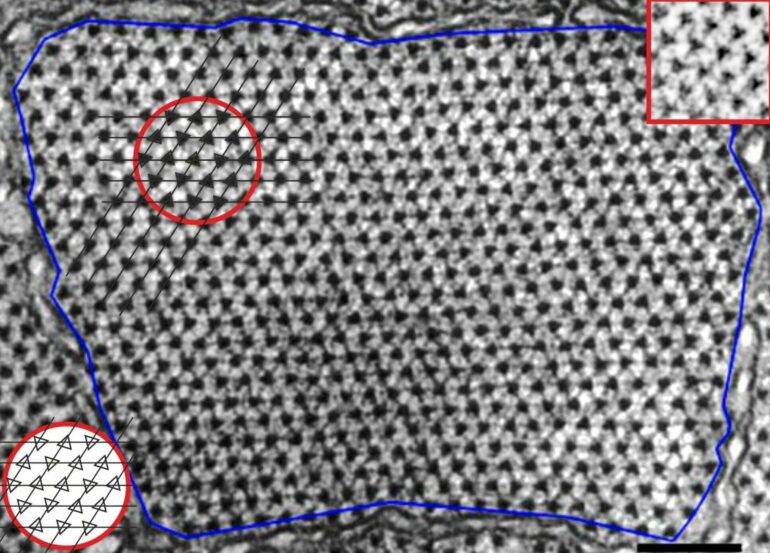
Myosin filaments pack together on a triangular array, and the myosin filaments on the array adopt one of two opposing orientations corresponding to two different orientations of the filament around its axis. The pattern of these “up” and “down” orientations of myosin filaments is irregular, but not entirely random.
This pattern of disorder, called the myosin superlattice, was first observed in the 1980s by Pradeep Luther and John Squire at Imperial following analysis of electron micrographs of muscle cross-sections They developed a set of empirical “rules” that described the arrangement, however until recently, the basis for this unique pattern was unknown.
More than 30 years later Rick Millane at the University of Canterbury in New Zealand noted that the distribution of the “up” and “down” orientations can be mapped to the “triangular Ising antiferromagnet” (TIA) model, a frustrated system from statistical physics. The researchers then conducted a detailed, quantitative analysis of the spatial distribution of the filament orientations observed in electron micrographs, which confirmed the equivalence to the TIA.
The model suggests that neighboring myosin filaments of the same orientation “repel” each other, causing them to arrange themselves so that filaments of opposite orientations are neighbors as much as is possible.
This interaction on a triangular array is the source of the frustration—if two neighboring filaments have opposite orientations, the third filament does not know which orientation to adopt. In the dining table analogy, it is not possible to avoid all disagreeable neighbors.
The mapping of the myosin filaments to the TIA model gives a deeper understanding of the source of the myosin array disorder and allows quantitative results such as the energetics of the adjacent myosin-myosin interactions to be estimated.
The model may also help to interpret X-ray diffraction data more accurately, allowing for improved imaging of muscle structure.
Fast and slow muscles
Not all muscle fibers show this disordered myosin superlattice structure. In some muscles, all myosin filaments have the same orientation—a pattern called a simple lattice structure.
Previous studies suggest that slow, fatigue-resistant muscles are often associated with the simple lattice structure, whereas fast, fatigable fibers are more often associated with the disordered superlattice. This indicates that the type of lattice may affect the mechanical properties of different types of muscle.
For example, the superlattice structure might allow more efficient sharing of actin binding sites by myosin heads, contributing to the greater force production of fast, fatigable fibers.
Prof Millane highlighted: “Further studies are needed to determine the effect of the different lattice structures on actin-myosin interactions and muscle behavior.”
Additionally, muscle structure varies across different species—with the simple lattice structure present in bony fish muscles, the slow muscles of sharks and the calf muscle of rats.
Dr. Luther also added: “Research investigating disordered myosin arrays in a wider variety of species may help us to understand the evolution of the muscle superlattice structure.”
More information:
Rick P. Millane et al, Geometric frustration in the myosin superlattice of vertebrate muscle, Journal of The Royal Society Interface (2021). DOI: 10.1098/rsif.2021.0585
Provided by
Imperial College London
Citation:
Complex model from physics observed for first time in vertebrate muscle (2022, April 21)