Proteins have been around a lot longer than we have—as building blocks of biological evolution, our existence depends on them. And now, researchers at the Georgia Institute of Technology are applying a 20th-century theoretical concept to study how proteins evolve, and it might lead to the answer of one of humanity’s oldest questions: How did we become us?
Inside a typical human cell are tens of thousands of proteins. We need so many because proteins are the skilled laborers of the cell with each one performing a specific job. Some lend firmness to muscle cells or neurons. Others bind to specific, targeted molecules, ferrying them to new locations. And there are others that activate the process of cell division and growth.
A protein’s specific function depends on its shape, and to achieve its functional shape—it’s native state—a protein folds. A protein begins its life as a long chain of amino acids, called a polypeptide. The sequence of amino acids determines how the protein chain will fold and form a complex, 3D structure that allow the protein to perform an intended task.
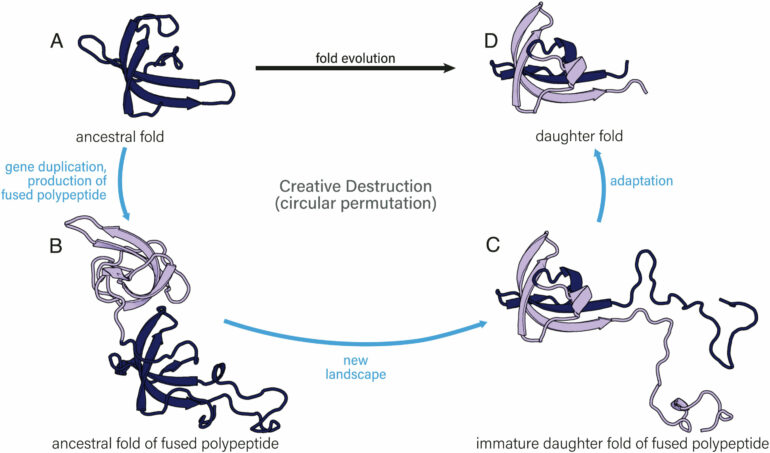
In the lab of Loren Williams, researchers are using “creative destruction” as a model for protein fold evolution and innovation. The term, coined by Austrian economist and political scientist Joseph Schumpeter in the 1940s, describes the deliberate dismantling of an established thing, like the wired telephone, to develop a new thing, like the smart phone.
“We have protein structures that have evolved over almost four billion years, and we don’t really understand where they came from or how they came to be what they are,” said Claudia Alvarez-Carreño, a postdoctoral researcher in the Williams lab, which is called the Center for the Origin of Life, or COOL. “It’s a very complex process forming these structures, and there are many hypotheses on how they could have emerged in early evolution.”
Out with the old, in with the new
Alvarez-Carreño is the lead author of the paper, “Creative Destruction: New Protein Folds from Old,” published recently in the journal Proceedings of the National Academy of Sciences, or PNAS. She and her co-authors (Williams, Rohan Gupta, and Anton Petrov) excavated the deepest evolutionary history found within the translation machinery—which resides within all cells in the ribosome and is the birthplace of all proteins.
The researchers provide evidence supporting the common origins of some of the simplest, oldest, and most common protein folds. It suggests a form of creative destruction at work, explaining how simple protein folds spawn more complex folds.
They discovered that once a protein can fold and achieve its 3D structure, when it is combined with another protein which has folded into a different 3D structure, that combination can easily become a new structure. “So maybe it’s not as difficult as we thought to go from one structure to another,” said Williams, professor in the School of Chemistry and Biochemistry. “And maybe this can explain the diversity of protein structures that we see today.”
In Schumpter’s creative destruction model, developing “daughter products” involves the destruction of ancestral products. “Daughter products can inherit features of ancestors but can in essence be different from them,” they write in the paper. In the smart phone example ancestral wired phones, computers, cameras, global positioning, and other technologies that are merged to create a daughter, i.e. the smart phone.
The daughter inherits many features of the ancestors. These features, which interact in specific ways in the daughter, create new functional niches that were not accessible, or even possible, in the ancestors.
“So, the creative destruction of protein folds might account for a lot of the diversity we see,” Williams said.
Molecular mergers
Ever since the simplest and most ancient protein folds emerged on Earth billions of years ago, the number of folds has expanded to form the universe of protein function we see in modern biology.
But the origins of protein folds and the evolutionary mechanisms at play pose central questions in biology that Williams and his team considered. For instance, how did protein folds arise, and what led to the diverse set of protein folds in contemporary biological systems, and why did nearly four billion years of fold evolution produce fewer than 2,000 distinct folds?
The researchers believe that creative destruction can be generalized to explain a lot of this.
In creative destruction, they explain, one open reading frame—the span of DNA sequence that encodes a protein —merges with another to produce a fused polypeptide. The merger forces these two ancestors into a new structure. The resulting polypeptide can achieve a form that was inaccessible to either of the independent ancestors, before the merger. But these new folds are not totally independent of the old. That is, a daughter fold inherits some things from the ancestral fold.
This, broadly speaking, is what Williams and his team observed, and they think their creative destruction model has some application in studying disease—proteins that fold improperly can impact the health of the cells and the human comprised of those cells.
“For example, we think this process is important in the biology of cancer—there are many, many proteins that have fused and, we believe have refolded, in cancers,” said Williams. “And there’s the world of protein aggregation diseases, like Parkinson’s or Alzheimers, and proteins that have not folded correctly, or have refolded.”
But right now, Williams and his team are most interested in how their creative destruction model helps them understand some of the deepest questions of our evolution.
“Like, where did we come from,” Williams said. “Creative destruction could help us understand where the proteins in our body came and how we came to be what we are.”
More information:
Claudia Alvarez-Carreño et al, Creative destruction: New protein folds from old, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2207897119
Provided by
Georgia Institute of Technology
Citation:
Creative destruction: Probing the evolution of proteins (2023, March 7)