During the replication of the SARS-CoV-2 virus, a long string of connected proteins is cleaved apart into individual proteins. This process is interrupted by an FDA-approved drug to treat COVID-19; however, the mechanistic details of this cleavage process are still unclear. Now, a team led by researchers at Penn State has produced the most detailed images to date of this process, revealing that these proteins are cleaved in a consistent order likely dictated by the structure of the protein string.
The results, published in a paper appearing in the Journal of Biological Chemistry, could support the development of more efficient drugs to treat COVID-19.
Retroviruses and many other RNA viruses—including SARS-CoV-2—translate their RNA genomes into a long string of connected proteins called a polyprotein. This polyprotein is later cleaved apart one at a time into individual, mature proteins by an enzyme called a protease.
This process is an essential step in virus replication and is thus an ideal target of antiviral drugs. For example, the drug Paxlovid, which has been approved for emergency use to treat mild-to-moderate COVID-19, contains a protease inhibitor called nirmatrelvir that interferes with this process in the SARS-CoV-2 virus within infected human cells.
“The SARS-COV-2 virus uses a protease called Mpro to cleave a polyprotein into 10 individual proteins in a specific order,” said Katsuhiko Murakami, professor of biochemistry and molecular biology at Penn State and an author of the paper.
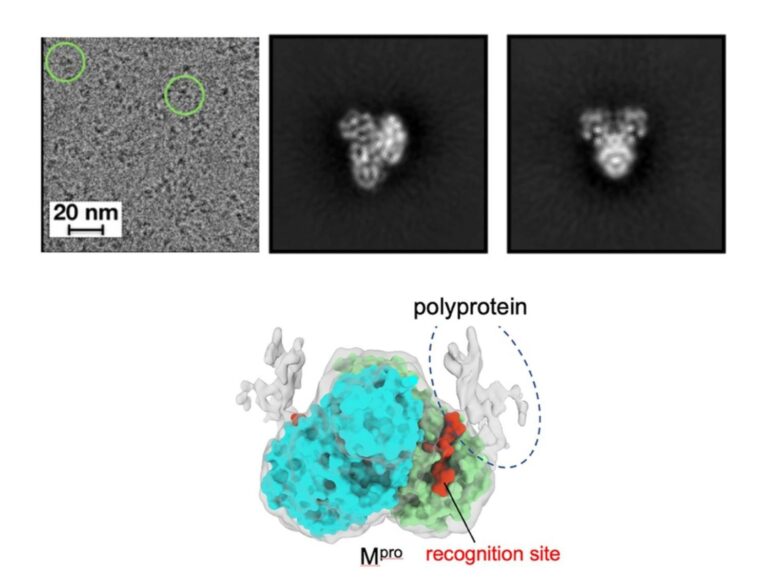
“But how this order is determined has remained unclear. Why does Mpro go to one recognition site first over the others? In this study, we used cryo-electron microscopy to produce high-resolution 3D visualizations of Mpro alone and in complex with the polyprotein. Better understanding how this cleavage process occurs could provide insights to optimize the binding of an antiviral drug or even reveal new ways to inhibit the process.”
Previous studies used an imaging technique called X-ray crystallography to investigate Mpro on its own or while it is attached to a very short segment of the polyprotein. However, the research team wanted to study Mpro in context, with a much more representative form of the polyprotein and at a higher resolution.
“A limitation with previous studies was they were using only a small peptide to mimic the polyprotein,” said Manju Narwal, postdoctoral researcher at Penn State and first author of the paper. “We wanted a clearer view of how Mpro approaches the various recognition sites on the polyprotein to see if there was something about Mpro or the polyprotein that dictates where it goes first.”
The researchers found that Mpro associates with the recognition site at the cleavage location but makes very little contact with the rest of the polyprotein. According to the researchers, this suggests that the polyprotein structure may dictate which site Mpro cleaves first.
“If Mpro was driving the process, selecting a preferred recognition site from all of the exposed possibilities, we would expect it to make additional contacts with the polyprotein for reasons related to energetics and stability,” said Narwal.
“Because we don’t see those additional contacts, we suspect that the polyprotein’s structure may instead dictate the cleavage order. For example, only a limited number of recognition sites within the polyprotein may be accessible to Mpro. When the first protein is cleaved by Mpro and separates from the rest of the polyprotein, the next preferred site is exposed. Then, once that one is cleaved, it exposes another, and so on.”
These new insights were possible because the researchers used cryo-electron microscopy (cryo-EM), an imaging technique that produces atomic resolution biomolecular structures.
“Because of the importance of Mpro, researchers from around the world quickly produced hundreds of images of the protease using X-ray crystallography, which is well-suited to investigating structures of smaller targets,” said Jean-Paul Armache, assistant professor of biochemistry and molecular biology at Penn State and an author of the paper.
“Cryo-EM is a technique more often applied to larger molecules, but Mpro, even when bound to the polyprotein, is still quite small, which can make it harder to determine the structure at high-resolution. Despite the small size, we were able to focus on Mpro using cryo-EM thanks to a combination of Manju’s careful sample preparation; the skill of our cryo-EM microscopist and co-author, Thomas Edwards; and modern data-processing techniques.”
The research team conducted its initial experiments with instruments at the Penn State Huck Institutes of the Life Sciences’ Cryo-EM Facility and followed up with additional imaging at the National Cancer Institute’s National Cryo-EM Facility in Frederick, Maryland.
“Although we determined a clear image of Mpro when bound to the polyprotein, the image of the polyprotein itself was less clear,” said Murakami. “We are working to visualize the entire complex and to focus in on other regions of the polyprotein. These insights will support future research on this critical step in the replication of many viruses and, we hope, will ultimately support the creation of efficient new antiviral drugs.”
More information:
Manju Narwal et al, SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction, Journal of Biological Chemistry (2023). DOI: 10.1016/j.jbc.2023.104697
Provided by
Pennsylvania State University
Citation:
Essential process for SARS-CoV-2 viral replication visualized (2023, May 30)