Imagine a country with a billion people, where every individual has different interests and different goals. You will never know their interests and goals until you ask them, but asking a billion people is not an easy task.
This is the same complex scenario that scientists face when we study bacteria. There are about a billion of them in a colony the size of the tip of a pencil, but when we look at the whole colony of bacteria, they all look the same and we assume that they will all fall victim to the same antibiotic. Not so, unfortunately.
Just like people, every single bacterium in a wound has its own goal. Some will thrive and multiply, others will migrate to other parts of the patient’s body, some will succumb to antibiotic treatment, and a few will lie low and go unnoticed.
These last ones are the troublemakers, because they are both able to survive antibiotics, and they are not detected by diagnostic antibiotic resistance testing. Finding these low-lying troublemakers among hundreds of billions of bacteria is like finding a needle in a haystack. They are very difficult to find, but they can render medical treatment useless.
“We know that these troublemakers, the needles in the haystack, exist because every now and then somebody jumps into the haystack and gets hurt by it. We also know that in some chronic bacterial infections, the haystack contains more than one needle,” says the senior researcher of a recent study on the problem, associate professor Christian Lentz.
Finding the bad bugs with fluorescence
Recently, researchers at UiT The Arctic University of Norway and CANS—Centre for new Antibacterial Strategies—found a clever new way to look at single bacteria and to find the antibacterial resistant ones, or the troublemakers, among them. The researchers can now even predict how those villains will behave and how dangerous they will become.
The findings are published in the journal Communications Biology.
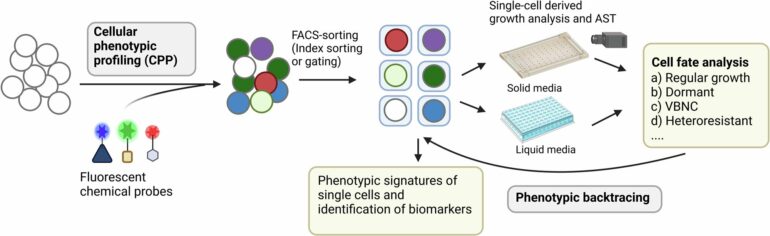
By combining fluorescent tags with the antibiotic Vancomycin used against the bacterium Staphylococcus aureus, the researchers were able to pinpoint single bacteria that look the same as the others but have the potential to do extra harm to patients suffering from Staphylococcus aureus infections.
“We are trying to paint ‘the needles’ with a fluorescent green color that can be easily spotted. For this we use special molecular ‘paints,’ for example antibiotics coupled to fluorescent dyes or other probes that tell us something about the needle-like molecular make-up of the bacterial cells.
“The combination of painting the cells in different colors, and correlating the color of the cells with their ability to survive antibiotics, allows us to predict if individual bacterial cells are more or less likely to be killed by antibiotics,” says Lentz.
Easier to choose the right antibiotic
Being able to know what types of antibiotic resistant troublemakers that hide within a bacterial colony can in the future prove vital in predicting the success or failure of a certain antibiotic treatment. This will make it easier to choose a more suitable antibiotic in the first place.
Hopefully, this will make us able to avoid unexplained antibiotic treatment failure where antibiotics that should work, according to diagnostics in the lab, fail to do so in the patient.
More information:
Jonathan Hira et al, Single-cell phenotypic profiling and backtracing exposes and predicts clinically relevant subpopulations in isogenic Staphylococcus aureus communities, Communications Biology (2024). DOI: 10.1038/s42003-024-06894-z
Provided by
UiT The Arctic University of Norway
Citation:
Fluorescence-activated cell sorting platform offers new way to look at single bacteria (2024, October 4)