by Soham Sengupta and Rajeev K. Azad
In the past few decades, there has been a rise in antibiotic-resistant infections, which are becoming an increasingly urgent public health concern. According to the World Health Organization, at least 700,000 people die each year due to antibiotic-resistant infections, and this number is only expected to increase as more and more bacteria become resistant to commonly used antibiotics.
Bacterial pathogens can disseminate infections or become resistant to drugs through a process called horizontal gene transfer (HGT). HGT allows bacteria to acquire new traits and adapt to new environments by exchanging genetic material with other species, even across the domains. Notwithstanding recent advances in the field, our understanding of bacterial evolution through HGT and dissemination of traits is limited.
Our recent study, published in the journal Open Biology, reveals new insights into HGT and its role in prokaryotic evolution. In this study, entitled “Reconstructing horizontal gene flow network to understand prokaryotic evolution,” we present a new approach for studying HGT, which allows identification of the donor and recipient organisms involved in gene transfer, particularly mobilization of not just alien but also long-term resident native genes across phyla, thus providing deeper insights into the complex network of genetic exchange between different bacteria taxa.
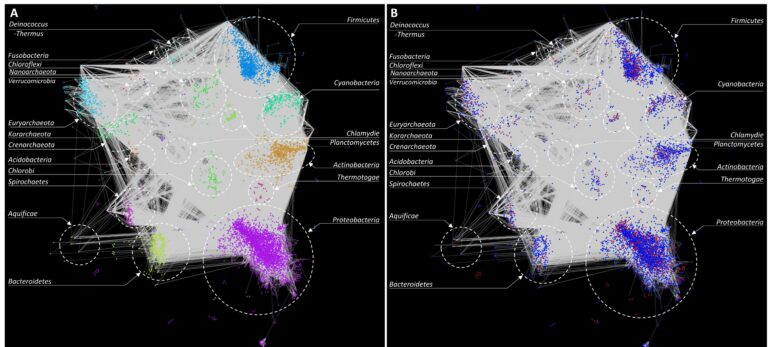
So how does this method work? Essentially, we used a combination of computational and statistical methods to reconstruct snapshots of the history of HGT events that potentially occurred between different bacterial taxa. We then used this information to create a “network” of HGT events, which allowed visualization of bacterial taxa that were involved in gene transfer and how frequently these events occurred.
The results of this study shone a new light on bacterial gene transfer. Our results reiterated that HGT is a widespread and an important mechanism of evolution, revealing numerous instances of these events even across distant phyla. This is made possible by certain genes or genomic segments possessing attributes that enable them to mobilize and integrate into the recipient genomes. This is pivotal for these bacteria to adapt and survive in different environments.
Most past studies have focused on cataloging alien genes in bacteria, without specifying the donor-recipient pair of a mobilized gene. By grouping genes originating from distinct sources and performing comparisons across all genomes, the direction of transfer of native genes could be established. This helped build the roadmap of gene transfer by constructing a gene flow network that specified donors and recipients, where possible. By combining multiple complementary methods, our approach effectively addressed the limitations of individual methods, resulting in a synergy of their strengths.
Our study also revealed that the gene transfer network is scale-free. The degree distribution of the network approximated a power-law distribution. This means that the connectivity between nodes (taxa) follows a scale-free or small-world pattern, which implies that the properties of a randomly sampled subset of the network would be similar to the properties of the entire network.
The HGT network presented in this paper has significant implications in advancing our understanding of bacterial evolution and adaptation. Gene mobilization and rapid dissemination of traits among bacterial pathogens pose significant challenges to public health, for example, the spread of antibiotic resistance facilitated by HGT. By understanding the mechanisms of HGT and the network of genetic exchange between different microorganisms, researchers can better comprehend the dissemination of antibiotic resistance through HGT and thus develop better strategies to combat it.
For example, by identifying the donor and recipient organisms involved in the transfer of resistance genes, researchers can focus their attention on the “perpetrators” of this health menace and plan strategies to prevent the spread of resistance. This knowledge could also help develop new approaches to controlling hotspots of infection and resistance, particularly focusing on such bacterial communities, thus preventing the spread of infections or antibiotic-resistant to the extent possible.
Our study could spur future research in this area, which is critical for understanding bacterial evolution in general and for developing effective solutions to public health problems caused by bacterial pathogens in particular.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information:
Soham Sengupta et al, Reconstructing horizontal gene flow network to understand prokaryotic evolution, Open Biology (2022). DOI: 10.1098/rsob.220169
Soham Sengupta, Ph.D., is a Bioinformatics Research Scientist at the St. Jude Children’s Research Hospital, Memphis, TN. This work has been published as part of his dissertation research at the University of North Texas, Denton, TX.
Rajeev K. Azad, Ph.D., is an Associate Professor in the Department of Biological Sciences and Department of Mathematics at the University of North Texas.
Citation:
Genes on the move: Mapping the pathways of horizontal gene transfer (2023, March 29)