LMU researchers have developed a method to extract more information from sequencing data. This will afford deeper insights into biology.
Higher organisms store their genetic material in the nuclei of cells as deoxyribonucleic acid (DNA). In a process called transcription, individual segments, the genes, are converted into messenger ribonucleic acids (mRNAs). Subsequently, the translation process produces proteins as the most important functional units.
Cells can turn various genes on or off. And diseases also have characteristic patterns of gene expression. This is why scientists try to sequence mRNA molecules so as to extract further information from them. The problem with established technologies is that information was often lost along the way.
A team led by Dr. Stefan Canzar from the Gene Center Munich has now presented Ladder-seq in Nature Biotechnology: a method that leads to better results. The scientists combine changes to the sequencing protocol with computer-assisted techniques. With the additional information, Canzar’s team is able, for example, to decode the function of regulatory units of neural stem cells in the brains of mice.
A jigsaw puzzle with additional information
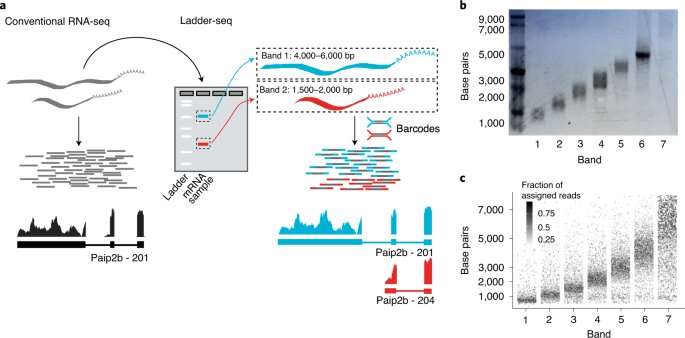
“In sequencing before now, the first step was to break down mRNA molecules, leaving you, figuratively speaking, with the jumbled pieces of a jigsaw puzzle,” explains Canzar. Modern devices then sequence these fragments in parallel, which saves time. With powerful computers, individual portions of the sequence are put back together again. Over the past 10 to 15 years, several methods have been further optimized. “Nevertheless, we know that information gets lost in the process that cannot be reconstructed,” says the LMU researcher.
Dr. Francisca Rojas Ringeling and Shounak Chakraborty from Canzar’s group have amended the usual protocol. Before the actual sequencing, researchers separate RNA molecules based on their length. The easiest way to do this is a technology called electrophoresis for separating molecules in an electric field. According to their size, mRNAs travel different distances. In the gel, they appear as a ladder, hence the name Ladder-seq. Only then do the researchers carry out fragmentation and sequencing. Algorithms use information about size to piece together individual parts of the puzzle with greater precision than previously possible.
Sample application in neurobiology
Using a sample application, the LMU researchers have demonstrated what Ladder-seq can accomplish. In the experiment, they looked at alternative splicing in the developing mouse brain. If you remove a chemical modification of RNA, the sequence of the RNA molecule itself changes. That is to say, you get alternative splicing. From the same gene, a different mRNA is produced—and consequently a different protein, which lacks certain structural domains. They no longer fulfill their biological roles in the brains of mice. Neural stem cells remain in this stage of development. And they no longer differentiate into nerve cells, because corresponding genes are dysregulated. The brains of affected rodents are strongly deformed and the creatures die shortly after birth. “Without the new method, we would never have been able to answer such questions with such precision and detail,” explains Canzar.
More information:
Francisca Rojas Ringeling et al, Partitioning RNAs by length improves transcriptome reconstruction from short-read RNA-seq data, Nature Biotechnology (2022). DOI: 10.1038/s41587-021-01136-7
Provided by
Ludwig Maximilian University of Munich
Citation:
Genome research: Finding the invisible (2022, January 11)