Researchers at the Texas A&M School of Veterinary Medicine & Biomedical Sciences (VMBS) and an interdisciplinary team of collaborators have uncovered new information about the history of cat evolution explaining how cats—including well-known species like lions, tigers, and domestic cats—evolved into different species, and shedding light on how different genetic changes in cats relate to survival abilities like the ability to smell prey.
By comparing genomes of several cat species, the project, published today in Nature Genetics, has helped researchers understand why cat genomes tend to have fewer complex genetic variations (such as rearrangements of DNA segments) than other mammal groups, like primates. It also revealed new insights into which parts of cat DNA are most likely to evolve rapidly and how they play a role in species differentiation.
“Our goal was to better understand how cats evolved and the genetic basis of the trait differences between cat species,” said Dr. Bill Murphy, a VMBS professor of veterinary integrative biosciences who specializes in cat evolution. “We wanted to take advantage of some new technologies that allow us to create more complete cat genomic maps.
“Our findings will open doors for people studying feline diseases, behavior, and conservation,” he said. “They’ll be working with a more complete understanding of the genetic differences that make each type of cat unique.”
Variations on a theme
Among the things the scientists were trying to better understand is why feline chromosomes—cellular structures containing the genetic information for traits like fur color, size, and sensory abilities—are more stable than in other mammal groups.
“We’ve known for a while now that cat chromosomes across species are very similar to each other,” Murphy said. “For example, the chromosomes of lions and domestic cats hardly differ at all. There appear to be far fewer duplications, rearrangements, and other types of variation than what are commonly found in great apes.”
In the primate order, this kind of genetic variation has led to the evolution of different species—including humans and great apes.
“The great ape genomes tend to break and rearrange, and even human genomes have very unstable regions,” Murphy said. “These variations may predispose certain individuals to have genetic conditions, like autism and other neurological disorders.”
The key to this variation between cats and apes, as Murphy found out, appears to be the frequency of something called segmental duplications—segments of DNA that are highly similar copies of other DNA segments found elsewhere in the genome.
“Primate genome researchers have been able to link these segmental duplications to chromosome rearrangements,” he said. The more segmental duplications you have in your DNA, the more likely the chromosomes are to rearrange, and so on.
“What we discovered by comparing a large number of cat species genomes is that cats have just a fraction of the segmental duplications found in other mammal groups—primates actually have seven times more of these duplications than cats. That’s a big difference, and now we believe we understand why cat genomes are more stable,” he said.
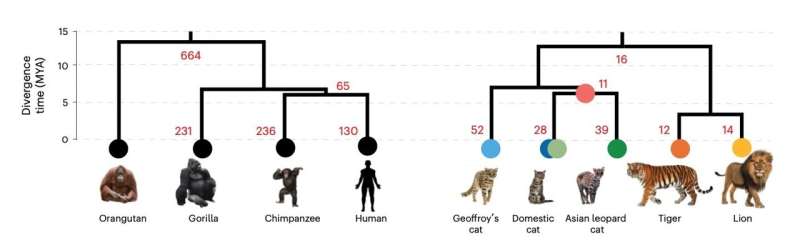
Graph comparing the number of certain genetic variations between primates and cats across an evolutionary timescale. © Dr. William Murphy, Texas A&M University, and colleagues
A needle in a (double) helix
While cats may not have as many large genetic rearrangements in their DNA, they still have plenty of differences. Through their research, Murphy and his colleagues now better understand which parts of cat DNA cause those variations, especially the variations that define speciation, or the differences between species.
“It turns out that there’s a large region on the center of the X chromosome where most of the genetic rearrangements are happening,” Murphy said. “In fact, there’s one specific repetitive element within this region called DXZ4 that evidence tells us is largely responsible for the genetic isolation of at least two cat species, the domestic and jungle cat.”
DXZ4 is what Murphy calls a satellite repeat—it’s not a typical gene that codes for a physical trait like fur color, but rather, it aids in the three-dimensional structure of the X chromosome and likely played an important role in cat speciation.
“We still don’t know the precise mechanism, but by comparing all these cat genomes, we can better measure the rate at which DXZ4 evolved in one species compared to all the others. What we learned is that DXZ4 is one of the most rapidly evolving parts of the cat genome; it’s evolving faster than 99.5% of the rest of the genome,” he explained.
“Because of the rate at which it mutates, we were able to demonstrate why DXZ4 is probably linked to speciation,” Murphy said.
Sniffing out elusive genes
Using new, highly detailed genome sequences, the team also uncovered clearer links between the number of olfactory genes, which govern scent detection in cats and variation in social behavior and how they relate to their surroundings.
“Since cats are predators who rely heavily on smell to detect their prey, their sense of smell is a pretty important part of who they are,” Murphy said. “Cats are a very diverse family, and we’ve always wanted to understand how genetic variation plays a role in different cat species’ ability to smell in their different environments.
“Lions and tigers have a pretty big difference between certain odorant genes involved in detecting pheromones, which are chemicals that different animals release into the environment to communicate information about identity, territory, or danger,” he continued.
“We think the large difference has to do with lions being very social animals living in family groups and tigers living a solitary lifestyle. Lions may have a reduced reliance on pheromones and other odorants because they’re constantly around other lions, reflected in the fewer genes of this type in their genomes,” he said.
Tigers, on the other hand, need to be able to smell prey across very large territories as well as find mates.
“Tigers, in general, have large olfactory and pheromone receptor repertoires,” Murphy explained. “We think this is directly tied to the size of their territories and the variety of environments in which they live.”
Domestic cats, on the other hand, appear to have lost a wide range of olfactory genes.
“If they don’t have to travel as far to find what they need because they’re living with people, it makes sense that natural selection wouldn’t preserve those genes,” he said.
Murphy shared that his favorite example from the project is the odorant receptors from the fishing cat, an aquatically adapted wild cat species living in Southeast Asia.
“We were able to show that fishing cats have retained many genes for detecting waterborne odorants, which is a pretty rare trait in terrestrial vertebrates,” he said. “All of the other cat species have lost these specific genes over time, but fishing cats still have them.”
This new information about olfactory genes in cats was made possible through a new approach to genome sequencing called trio binning, which allows researchers to sequence the most difficult regions of a genome.
This new technology also makes separating maternal and paternal DNA much easier.
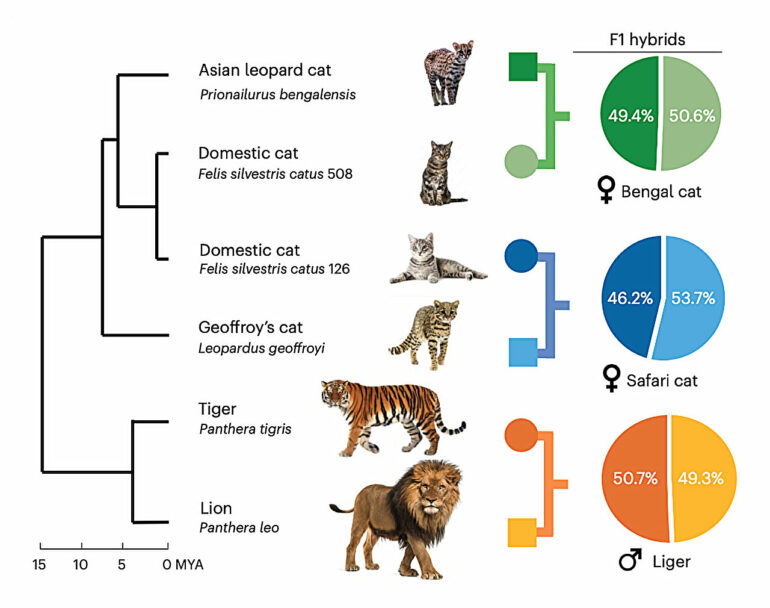
“With trio binning, you can now take DNA from an F1 hybrid—an animal whose DNA is split 50-50 between parents of different species—and cleanly separate the maternal and paternal DNA, giving you two complete sets of DNA, one for each parent species,” Murphy said. “The process is much simpler, and the results are more complete.”
Filling in the blanks
One of the most important conclusions from the project is that cat species may be similar in many ways, but their differences matter.
“These differences are showing us how these animals are perfectly suited for their natural environments,” Murphy said. “They’re not interchangeable, and that’s valuable information for conservationists and others working to preserve or restore species in their natural habitats.
“For example, you can’t assume that tigers from Sumatra and Siberia are the same,” he said. “Their environments are wildly different, and those tiger populations have likely developed specialized genetic adaptations to help them survive in these very different places.”
It’s also important for scientists to realize that the sections of genomes that are the most difficult to assemble may just be the key to understanding crucial bodily systems like immunity and reproduction.
“Olfactory genes aren’t the only ones that have been challenging to sequence and study. Scientists have also struggled to sequence immune and reproductive genes, so previous studies are missing this kind of information. Imagine trying to study a genetic condition in cats, humans, or any species, for that matter, without having all the pieces; this is why assembling complete genomes matters,” Murphy said.
For now, Murphy and his team will continue applying the most advanced genome sequencing and assembly technologies to cat genomes in order to fill in as much information as possible about the world of cats.
The study was conceptualized by Bill Murphy—VMBS professor of veterinary integrative biosciences at Texas A&M and Wes Warren—professor of genomics in the Bond Life Sciences Center at the University of Missouri. Additional collaborations involved researchers from the University of Washington, University College Dublin, the Institute for Systems Biology in Seattle, Louisiana State University and the Guy Harvey Oceanographic Center.
More information:
Kevin R. Bredemeyer et al, Single-haplotype comparative genomics provides insights into lineage-specific structural variation during cat evolution, Nature Genetics (2023). DOI: 10.1038/s41588-023-01548-y
Provided by
Texas A&M University
Citation:
Genome sequencing project reveals new secrets about cat evolution (2023, November 2)