Researchers have imaged a major component in conjugation—the process bacteria use to share DNA with each other.
During conjugation, bacteria can exchange genetic information in the form of special pieces of DNA. These include genes that help them resist attacks from common antimicrobial drugs, making many illnesses caused by these bacteria resistant to treatment.
Better understanding conjugation could therefore allow scientists to find ways to stop the process and reduce the spread of antimicrobial resistance.
Although conjugation has been studied since the 1940s, the atomic details underlying the mechanism were unknown. Now, researchers from Imperial College London and Queen Mary University of London have imaged a key part of the process for the first time.
The results, published in Nature Communications, are an important step forward in understanding how bacteria acquire drug resistance—and potentially how to stop them doing so.
Lead researcher Dr. Tiago Costa, from the MRC Center for Molecular Bacteriology and Infection and Department of Life Sciences at Imperial, said: “Our findings set the stage for an acceleration of our understanding of gene transfer conferring antimicrobial resistance among bacteria. Importantly, deciphering the mechanistic details of gene transfer could provide us with targets that can be exploited to suppress antibiotic resistance.”
The team studied E. coli bacteria. When an E. coli bacterium wants to initiate conjugation with another bacterium, it uses a type IV secretion system (T4SS) – a structure that reaches out and penetrates the other bacterium.
Many types of bacteria use the T4SS for a range of processes, including injecting toxic proteins into our own cells during infection, or injecting toxins into rival bacterial cells to kill them when competing for resources.
During conjugation, it’s not toxic proteins that are transferred, but segments of free-ranging DNA that confer an advantage to bacteria—including antimicrobial resistance. These DNA segments can even be transferred to different species of bacteria, spreading drug resistance to new pathogens.
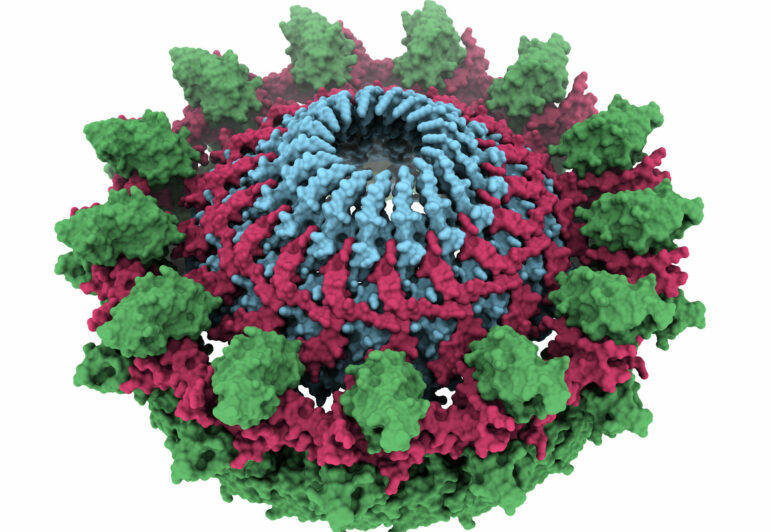
The T4SS is composed of three parts. The team were able to image the part called the outer-membrane core complex, which anchors the T4SS to the bacterium that is sharing its genes. From this anchor, the bacterium sends out a pilus—a long strand that grabs hold of the neighboring bacterium to which the DNA is to be transferred.
The images, obtained using a technique called cryo-electron microscopy, revealed that the outer-membrane core complex is made up of two concentric rings of proteins with very flexible connectors. This structure is highly flexible, which researchers believe help with the extension and retraction of the pilus as it reaches out to the other bacterium.
First author of the paper Ms Himani Amin, also from the Department of Life Sciences, said: “Our structure reveals fascinating new insights into the outer membrane core complex by displaying remarkable protein organization within. This will pave the way towards a better understanding of gene transfer amongst bacterial population and could enable researchers to develop preventative therapeutics.”
The team now want to image the rest of the conjugation T4SS in E. coli, revealing its full functionality in sufficient detail to show scientists how the process could be disrupted, reducing the transfer of antimicrobial resistance.
“Architecture of the outer-membrane core complex from a conjugative type IV secretion system” by Himani Amin, Aravindan Ilangovan and Tiago R. D. Costa is published 25 November 2021 in Nature Communications.
More information:
Himani Amin et al, Architecture of the outer-membrane core complex from a conjugative type IV secretion system, Nature Communications (2021). DOI: 10.1038/s41467-021-27178-8
Provided by
Imperial College London
Citation:
Key step in how bacteria acquire drug resistance revealed (2021, November 25)