When genes are activated and expressed, they show patterns in cells that are similar in type and function across tissues and organs. Discovering these patterns improves our understanding of cells—which has implications for unveiling disease mechanisms.
The advent of spatial transcriptomics technologies has allowed researchers to observe gene expression in their spatial context across entire tissue samples. But new computational methods are needed to make sense of this data and help identify and understand these gene expression patterns.
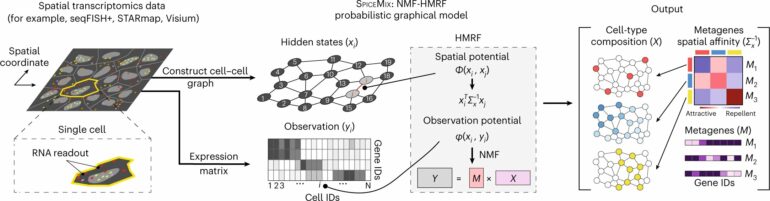
A research team led by Jian Ma, the Ray and Stephanie Lane Professor of Computational Biology in Carnegie Mellon University’s School of Computer Science, has developed a machine learning tool to fill this gap. Their paper on the method, called SPICEMIX, appeared as the cover story in the most recent issue of Nature Genetics.
SPICEMIX helps researchers untangle the role different spatial patterns play in the overall gene expression of cells in complex tissues like the brain. It does so by representing each pattern with spatial metagenes—groups of genes that may be connected to a specific biological process and can display smooth or sporadic patterns across tissue.
The team, which included Ma; Benjamin Chidester, a project scientist in the Computational Biology Department; and Ph.D. students Tianming Zhou and Shahul Alam, used SPICEMIX to analyze spatial transcriptomics data from brain regions in mice and humans. They leveraged the unique capabilities of SPICEMIX to uncover the landscape of the brain’s cell types and spatial patterns.
“We were inspired by cooking when we chose the name,” Chidester said. “You can make all sorts of different flavors with the same set of spices. Cells may work in a similar way. They may use a common set of biological processes, but the specific combination they use gives them their unique identity.”
When applied to brain tissues, SPICEMIX identified spatial patterns of cell types in the brain more accurately than other methods. It also uncovered new expression patterns of brain cell types through the learned spatial metagenes.
“These findings may help us paint a more complete picture of the complexity of brain cell types,” Zhou said.
The number of studies using spatial transcriptomics technologies is growing rapidly, and SPICEMIX can help researchers make the most of this high-volume, high-dimensional data.
“Our method has the potential to advance spatial transcriptomics research and contribute to a deeper understanding of both basic biology and disease progression in complex tissues,” Ma said.
More information:
Benjamin Chidester et al, SpiceMix enables integrative single-cell spatial modeling of cell identity, Nature Genetics (2023). DOI: 10.1038/s41588-022-01256-z
Provided by
Carnegie Mellon University
Citation:
Machine learning method improves cell identity understanding (2023, January 13)