A new computational framework created by Oak Ridge National Laboratory researchers is accelerating their understanding of who’s in, who’s out, who’s hot and who’s not in the soil microbiome, where fungi often act as bodyguards for plants, keeping friends close and foes at bay.
The research is shining a light on specialized fungal metabolites, small molecules produced by fungi that can help or harm plants and other organisms. These metabolites can, for instance, help bioenergy plants like the poplar tree thrive in harsh growing conditions and store more carbon belowground. Fungal metabolites are also used by humans to make everything from food additives such as citric acid to drugs to fight bacterial infections and cancer, and as pesticides and herbicides for agriculture.
The gene clusters in fungi that produce specialized metabolites are usually silent in standard laboratory cultures until signaled by external compounds or environmental conditions. ORNL scientists wanted to analyze the interactions of compounds like lipids and chitins on metabolite production in fungi to improve the accumulation of these natural products.
Using a data-driven approach described in the journal PNAS Nexus, ORNL scientists were able to predict which substances would best stimulate fungal metabolites, then validated the findings in the lab using cultures of the mold Aspergillus fumigatus, mass spectrometry analysis and comparison to published datasets. The approach greatly speeds up the painstaking process typically deployed to identify, extract and characterize specialized metabolites.
The modeling framework leverages graph theory, a machine learning approach that analyzes the complex relationships in processes such as the interactions of microbes with each other and with plants.
“By using graph theory, we can better identify the chemical signals that trigger certain metabolites in fungi, and we can also use it to narrow down which fungal species are important to analyze,” said Muralikrishnan (Murali) Gopalakrishnan Meena, computational scientist at ORNL’s National Center for Computational Sciences.
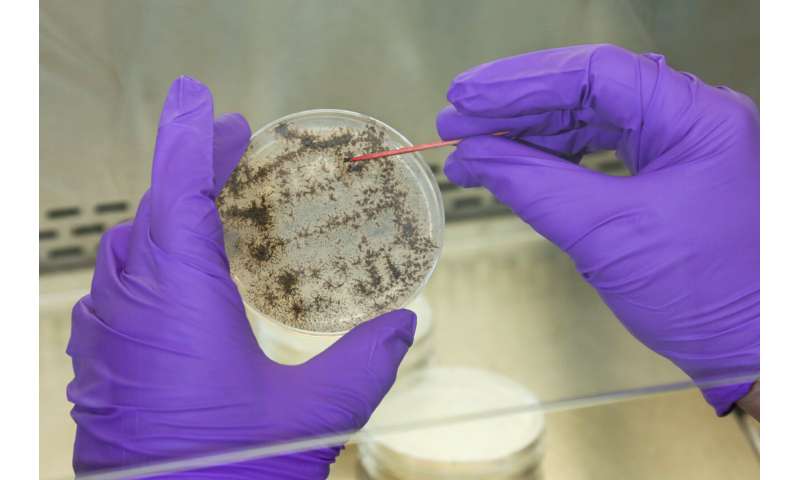
Tomás Rush analyzes a fungus culture as part of ORNL’s research for the Plant-Microbe Interfaces Scientific Focus Area. © Genevieve Martin/ORNL, U.S. Dept. of Energy
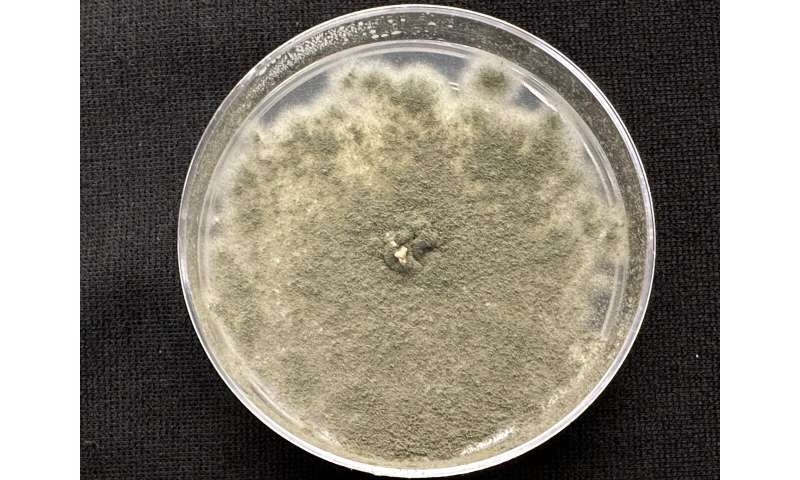
Shown here is a culture of the fungus Aspergillus fumigatus, one of several microbes being studied at ORNL for their impact on bioenergy crops such as the poplar tree. © Tomás Rush/ORNL, U.S. Dept. of Energy
The research is part of the Plant-Microbe Interfaces Scientific Focus Area at ORNL, a project that aims to better understand the mutually beneficial relationships between plants and microbes in the plant root environment known as the rhizosphere. That information can then be used to address challenges related to bioenergy, environmental remediation and soil carbon storage.
The SFA is largely focused on poplar—a key biomass crop being studied at ORNL for bioenergy production.
“We want to understand the effects of metabolites, organisms and compounds on poplar—its roots, stems, leaves, the entire microbiome,” said Tomás Rush, a mycologist in ORNL’s Biosciences Division who co-led the project.
Some fungi like A. fumigatus, a microbe important to carbon cycling found in the poplar rhizosphere, are good at identifying both threats and benefactors in the soil microbiome that can affect plant hosts, Rush said.
Fungi ‘drop the rope’ for plant-friendly microbes
“Aspergillus appears to be letting some microbes get close to the rhizosphere while keeping others out,” Rush said. “You have tons of microbes in the soil, but why is it we’re only finding a few in the poplar rhizosphere all the time? I think of it as a nightclub where the bouncers will keep some people out while dropping the rope at the entrance to let others in. That’s what Aspergillus is doing, acting as bodyguards to let some microbes get close to the plant while walling others off. “
This gatekeeping relationship was described more in-depth in a publication in mSystems, which explored the role of a microbial-derived signaling molecule in a beneficial plant-fungus symbiosis.
In the past year, Rush and his colleagues have collected and identified more than 1,500 fungi—isolated from the poplar symbiotic environment—using experimental and computational approaches, doubling the previous database.
“There’s already a lot of computational and bioinformatics work being done as part of the Plant-Microbes SFA,” Gopalakrishnan Meena said. “With this project, we bring in network science tools to classify these interactions with the fresh perspective of someone who is not a biologist, but is helped to understand the issue by colleagues who are. We can use the same data-driven, AI-type tools on many complex science challenges.”
The project used resources of the Oak Ridge Leadership Computing Facility, a DOE Office of Science user facility at ORNL, such as the JupyterHub platform supporting multi-user, data-driven analysis with seamless access of data and codes at a single hub.
The researchers first started working together after Gopalakrishnan Meena attended a poster session at which Rush was presenting his fungal-plant work when they were both postdoctoral researchers at ORNL. Gopalakrishnan Meena was looking for new applications for his machine learning expertise, which had mostly been applied at that point in his career to model turbulence in the atmosphere and in oceans.

ORNL’s Murali Gopalakrishnan Meena developed a computational framework to better analyze the compounds and environmental stresses affecting fungal metabolite production. © Carlos Jones/ORNL, U.S. Dept. of Energy
“ORNL and the PMI project in particular are good at fostering early career scientists like Murali and me to collaborate and explore ideas with new, interdisciplinary approaches,” Rush said.
“It’s exciting to see our early career researchers leverage our unique capabilities at ORNL to find new ways to address hard problems. There are thousands of metabolites working at the interface between plants and microbes. We need to know when they are being used and what they are doing,” said Mitchel Doktycz, ORNL Corporate Fellow and head of the lab’s Bioimaging and Analytics Section, who leads the PMI SFA.
Other ORNL scientists on the project include Matthew Lane and Armin Geiger, both also students at the Bredesen Center for Interdisciplinary Research and Graduate Education at the University of Tennessee; Joanna Tannous, Alyssa Carrell, Paul Abraham, Richard Giannone and Daniel Jacobson. Two former ORNL scientists who worked on the project are Jesse Labbé, now at TekHolding, and David Kainer, now at the Australian Research Council Centre of Excellence for Plant Success in Nature and Agriculture. Jean-Michel Ané and Nancy Keller of the University of Wisconsin-Madison also contributed.
More information:
Muralikrishnan Gopalakrishnan Meena et al, A glimpse into the fungal metabolomic abyss: Novel network analysis reveals relationships between exogenous compounds and their outputs, PNAS Nexus (2023). DOI: 10.1093/pnasnexus/pgad322
Provided by
Oak Ridge National Laboratory
Citation:
Modeling framework finds fungal ‘bouncers’ patrol plant-microbe relationship (2024, January 17)



