A innovative molecular “cut and sew” process by University of Dundee scientists has allowed the design of a research tool that will accelerate drug design for diseases for which no other options exist, including cancer.
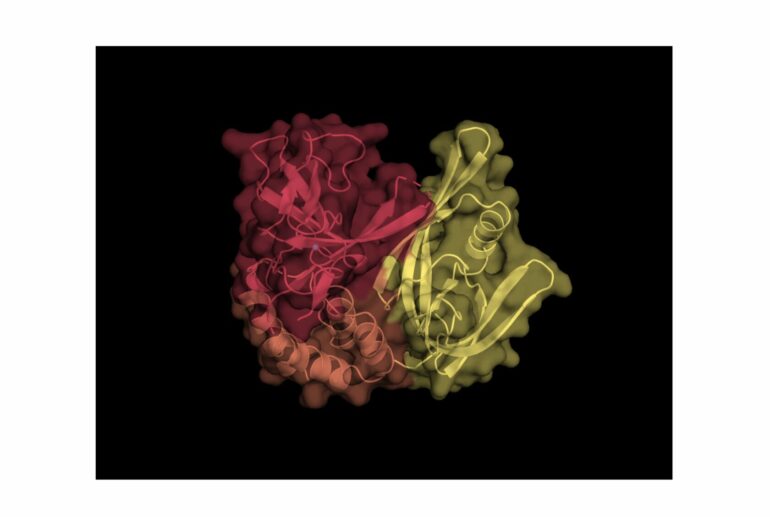
Experts from the University’s Center for Targeted Protein Degradation have custom-built a new version of a protein that is commonly hijacked by molecules known as “protein degraders.” The process removes unwanted bits of the protein before the remaining parts are stitched together, allowing researchers to isolate the protein and work with it more efficiently in the laboratory.
This pioneering work has been published in the journal, Nature Communications.
Protein degraders are heralding a revolution in drug discovery, with more than 50 drugs of this type being currently tested in clinical trials. These types of molecules work in a way that is fundamentally different from the way conventional drugs work.
Instead of just blocking disease-causing proteins, degrader drugs recruit or hijack them to stick to an important protein called ubiquitin E3 ligase, which works as part of the cellular protein recycling machinery. This recruitment is fundamental to how the drug works and leads to the disease-causing protein being destroyed and completely removed from inside the cell.
The majority of the drugs currently dosed in patients that work this way hijack a single E3 ligase called Cereblon (CRBN). Until now, approaches to design safer and more drug-like molecules and understanding how they recruit disease-causing proteins to CRBN has been slow and inefficient.
However, the Dundee team, led by structural biologists Dr. Alena Kroupova and Dr. David Zollman, in the group of Professor Alessio Ciulli, have devised an ingenious approach to design a new variant of the protein that they have called “CRBN-midi.”
This involves removing unwanted bits of the protein and stitching the remaining parts together. This improves the ability to isolate the protein and work with it in the laboratory.
Dr. Kroupova, first author of the study, said, “Our variant of the CRBN protein is more soluble and more easily crystallizable than the parent protein, while behaving closely to it and maintaining its biological characteristics.
“This makes it highly suitable for many of the experiments and processes today widely used in the field of targeted protein degradation.
Dr. Zollman, co-corresponding author on the study, added, “With TPD revolutionizing the world of drug design, this breakthrough will further help accelerate the generation of new degrader drugs that work through this protein, which will ultimately benefit patients living with some of the most pressing health challenges of our time.”
“We have made our CRBN-midi reagent widely available to the scientific community with no restrictions and no strings attached, via the Addgene portal. This has already been highly popular since we first disclosed it as a preprint in January 2024,” said Professor Ciulli.
“Many research groups around the world, including from academia and the biopharma industry, are already using it for their research and drug discovery programs.”
More information:
Alena Kroupova et al, Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders, Nature Communications (2024). DOI: 10.1038/s41467-024-52871-9
Provided by
University of Dundee
Citation:
Molecular ‘cut and sew’ process could accelerate drug design (2024, October 18)