Some of the toughest challenges in treating disease are presented by “undruggable” proteins whose structures and roles in disease are known but are seemingly unable to be targeted by drugs that will bind to them. Researchers at KAUST have now shown that the molecular motion of many “undruggable” proteins can in fact expose sites at which drugs could bind.
Their study is published in Nature Communications.
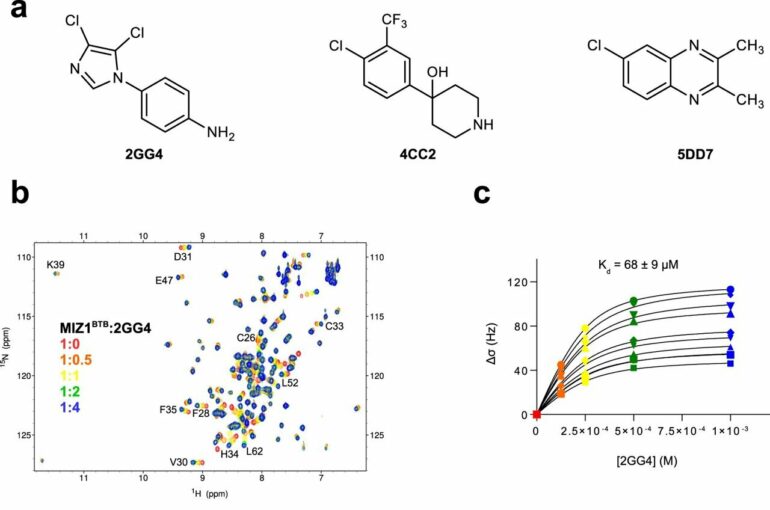
The research is focused on a particular molecular region, called the BTB domain, that is known to be a critical part of more than 350 proteins. It allows proteins to bind to other proteins to influence complex genetic and molecular signaling processes central to the activities of many cells.
More than 80 known BTB-containing proteins are transcription factors that control the activities of genes, a role that means many of them are implicated in cancer. As the BTB domain has proved difficult to target with drugs, these cancers are often fatal.
The team at KAUST, together with colleagues at the University of Michigan in the U.S., conducted a detailed analysis of the molecular motions of BTB domains in three proteins involved in cancer.
The results uncovered the role of molecular motion in influencing the ability of small molecules, collectively known as ligands, to bind to the BTB domain. This revealed cryptic binding sites—dynamic regions of BTB domains that appear available to bind to ligands, unlike the static structures.
“This means that some seemingly undruggable target proteins can now be reconsidered, with the firm hope of identifying novel lead compounds for anticancer drug development,” says Łukasz Jaremko of the KAUST team. “The hero of our study, called the MIZ1 protein, is linked to c-MYC, the oncogene cancer-causing gene of over 70% of cancers, and can now be targeted for drug discovery campaigns.”
The researchers were surprised to discover how important the movement of proteins could be in controlling ligand binding sites, while acknowledging it seems logical in retrospect.
First author Vladlena Kharchenko, a former KAUST Ph.D. student and now a postdoctoral fellow at Albert Einstein College of Medicine in the U.S., says the next challenge is to fully understand the mechanisms that allow molecular movements to make cryptic binding sites so hard to detect and interact with.
“We also want to find these sites in other proteins, to advance the drug discovery process for many other currently undruggable proteins and ultimately give new hope for treating currently incurable diseases, including many forms of cancer,” Kharchenko concludes.
More information:
Vladlena Kharchenko et al, Increased slow dynamics defines ligandability of BTB domains, Nature Communications (2022). DOI: 10.1038/s41467-022-34599-6
Provided by
King Abdullah University of Science and Technology
Citation:
Molecular motion of proteins reveals previously unseen binding sites that could be targets for new drug molecules (2022, November 21)