For many diseases, the tools of medicine are reaching their limits. CRISPR technologies open new avenues for diagnostics and therapies, although the natural source of CRISPR remains largely untapped. Scientists from the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, a joint venture of the Helmholtz Centre for Infection Research (HZI) in Braunschweig and the Julius Maximilian University (JMU) Würzburg, now demonstrate a new way to study CRISPR systems that can accelerate their conversion into tools. Their approach was published today in the journal Molecular Cell.
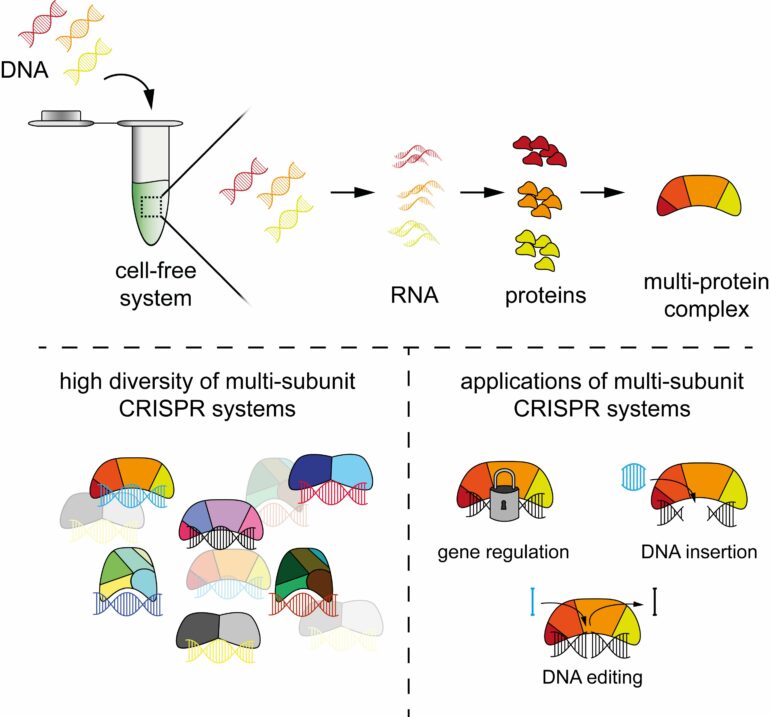
CRISPR technologies are tools used for genome editing, among other things. Often referred to as “gene scissors,” these tools derive from immune systems that protect bacteria from invading viruses. When applied, CRISPR technologies are often based on single proteins such as Cas9 or Cas12 and a ribonucleic acid (RNA) component that directs the protein to its target sequence. However, three-quarters of the bacterial immune systems involve not one but multiple proteins forming a complex.
Unique functions in the defense against viruses
These multi-protein complexes perform unique functions in defending against viruses and could lead to new applications for humans as well. Yet the large number of proteins makes it difficult to complete the steps necessary for characterisation.
“These challenges have limited our fundamental understanding of CRISPR systems involving multiple proteins and their use as technologies,” says Franziska Wimmer, one of the two first authors of the study published today in the journal Molecular Cell. “Against this background, we now present a promising new approach,” adds the HIRI Ph.D. student.
The new approach is based on cell-free systems (abbreviated as TXTL)—a liquid that can produce RNAs and proteins from added DNA. In combination with other technologies such as next-generation sequencing, TXTL can be used to quickly produce protein complexes and study their functions. The entire process requires less than a day to complete.
“Standard methods that use purified proteins or cell-based samples are extremely slow, making it hard to study most CRISPR systems found in nature,” says Dr. Ioannis Mougiakos, second first author of the current study. “In comparison, our new approach, called PAM-DETECT, is a real boost for CRISPR research. It can radically advance our efforts in the future.”
“We hope that PAM-DETECT will be widely adopted by the CRISPR community, and we’ve already started to collaborate with multiple groups applying the technique,” says Chase Beisel. The corresponding author of the study is a professor at JMU and head of the RNA Synthetic Biology Department at HIRI, which also includes Wimmer and Mougiakos.
PAM-DETECT determines which DNA sequences can be targeted with a CRISPR system. Using this technique, HIRI researchers were able to characterize a large set of systems involving protein complexes. They also applied this technique to study CRISPR transposons, which can be harnessed to insert large pieces of DNA. The findings of the current study could lead to new CRISPR technologies for diagnostics, gene editing, and gene regulation.
More information:
Franziska Wimmer et al, Rapid cell-free characterization of multi-subunit CRISPR effectors and transposons, Molecular Cell (2022). DOI: 10.1016/j.molcel.2022.01.026
Provided by
Helmholtz Association of German Research Centres
Citation:
New approach enables faster characterization of CRISPR immune systems (2022, February 25)