The products of photosynthesis are easy to point out. Plants, algae and cyanobacteria create the air we breathe and the fuel for food webs as they turn carbon dioxide and water into oxygen and sugars. How photosynthesis works, though, is much harder to pin down.
Photosynthesis appears to have a lot in common with a Rube Goldberg machine: surprising dependencies, intricate connections, and many, many moving parts. A lot of these moving parts are proteins—amino acid structures that harvest light, transport molecules and regulate reactions. Identifying these proteins and then figuring out what they are doing is difficult.
Proteins are tiny, invisible to even the most powerful light microscope, and fickle, often hiding their function from even the most determined scientist.
Work appearing in Nature Communications takes on the challenge of understanding one of these proteins—and its role in photosynthesis—in the cyanobacterium Synechocystis.
Through an array of experiments at universities and national user facilities, researchers found that this protein detects iron and fine-tunes energy production, allowing cyanobacteria to quickly adapt to their environment.
“The protein we studied is a very interesting regulator that can make the link between photosynthesis, respiration and high-iron homeostasis,” said Nicolas Grosjean, the first author of this paper. Grosjean is a research scientist at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab).
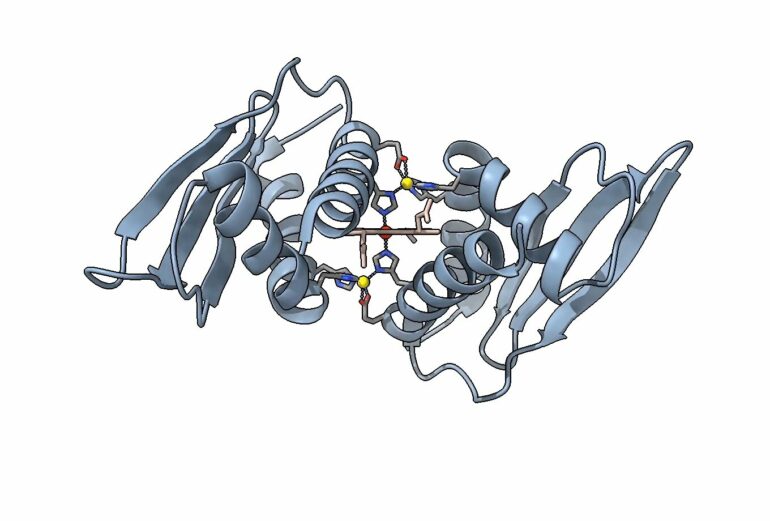
Structurally, this protein is unique, as well. “The heme-metal site that we found in the crystal structure, as far as we know, is a type of metallic protein that no one’s ever had a structure of before,” said Crysten Blaby-Haas, a research scientist at the JGI and Molecular Foundry, and the study’s senior author.
In other proteins, two histidine residues can be found to sandwich an iron-containing heme molecule. In the protein structure Grosjean and Blaby-Haas studied, that sandwich has an additional outer layer—two zinc ions, a component they’ve named a zinc-mirror heme site.
To get so much detail on this structure, Grosjean and Blaby-Haas collaborated with a range of researchers across the Albert Einstein College of Medicine, Washington University in St. Louis and the University of California at Berkeley, as well as other DOE user facilities—Brookhaven National Laboratory’s National Synchrotron Light Source II, and SLAC National Accelerator Laboratory’s Stanford Synchrotron Radiation Light Source.
“DOE has some amazing resources and people—not just the instrumentation—but the experts we were able to work with on this project to ask questions about structure and function,” Blaby-Haas said.
Looking at blueprints in high-throughput
This work builds on many other efforts to understand the Rube Goldberg machine of photosynthesis. Historically, this is a machine that has evaded direct experiments, so researchers have studied the blueprint plans that give rise to the protein parts of that machine—photosynthesizers’ DNA and RNA. These sequences provide a list of machine parts, and an idea of their groupings.
One pivotal effort along these lines came from UC Berkeley professor and longtime JGI collaborator Sabeeha Merchant’s group. In 2011, they inventoried a swath of blueprints to find parts that photosynthesizers share. The end result was a list of genes, termed The GreenCut2 Resource. It details 597 genes coding for proteins that are unique to photosynthesizing organisms.
“And perhaps surprising to a lot of people—a lot of the genes on that list—we have no idea what they’re doing,” Blaby-Haas said. Yet these genes are key to photosynthesis. Understanding them could unlock new ways of cycling carbon, optimizing crops and creating bioproducts.
So Blaby-Haas and her colleague Ian Blaby, a co-author of the new study, set out to tackle some of these genes, expressing and purifying the range of proteins these genes produced, to identify examples that would work well for in vitro and in vivo experiments.
In a project led by Blaby, through what is now the Community Science Program Functional Genomics proposal call, the JGI synthesized hundreds of genes representing a portion of GreenCut genes with unknown or uncertain function. These genes would allow the team to express this library of encoded proteins at high throughput. “The JGI synthesis project was the spark that started it all,” Blaby-Haas said.
A more apt name for a Jane Doe
After screening hundreds of GreenCut2 proteins, Blaby-Haas emerged with several ideas of proteins that seemed interesting and feasible to look into. These candidates would work well in further experiments, so she could glean information about their specific function, filling in our understanding of photosynthesis.
One initial target was a family of proteins that appeared in plants and algae and contained a similar region— a domain of unknown function, DUF2470.
In the world of proteins, a domain of unknown function is a bit of a Jane Doe—anonymous, mysterious, ubiquitous. Blaby-Haas and her team set out on a project that would eventually let them put a face to a name for this domain of unknown function.
First, they looked across the tree of life, doing a very complex CTRL+F search for this domain, looking for context clues that might point them toward the domain’s function. They found many instances of this domain, often bound to other protein structures. Repeatedly, this search surfaced another important partner for this protein—an iron-bound molecule called heme.
Photosynthesizers depend on iron and heme for the light-dependent reactions of photosynthesis, so this was an exciting partner. The picture of this Jane Doe was coming into focus. Blaby-Haas and her team hypothesized that DUF2470 was key to iron metabolism, and renamed it to “Domain Related to Iron,” or DRI for short.
A kaleidoscope of structure-function data
To study DRI experimentally, Blaby-Haas, Grosjean and their team used a model organism that produces this domain as a stand-alone protein, the cyanobacterium Synechocystis. Working with Synechocystis also opened up options for studying modified versions of this protein domain.
In an array of computational, in vitro and in vivo experiments, they looked at how this domain binds iron, and how that relates to the way Synechocystis cycles carbon.
These experiments leveraged the DOE Office of Science facilities in complementary ways to investigate this protein domain. Scientists at Brookhaven National Laboratory’s NSLS-II used multiple X-ray techniques combined with advanced computational simulations to take pictures of the protein with and without its heme partner, revealing that the protein contains zinc atoms mirroring one another on either side of the heme.
Scientists at Stanford’s SSRL were able to provide crucial data supporting the presence of the zinc-mirror heme site—the exciting structural component that sets this molecule apart.
To better understand how the revealed structure of the protein related to its function inside the cell, Grosjean used the JGI-developed guideRNA and Sequence Extraction Tool (gRNA-SeqRET) to produce knock-out mutants for experiments. When the cyanobacterium loses this protein, energy metabolism is disrupted.
These data create a multilayered understanding of this Domain Related to Iron, an effort that was interdisciplinary and collaborative. “All the user facilities that we worked with are cutting edge in terms of analysis and the fascinating results that we managed to compile,” Grosjean said.
With this breadth of results, this team has recast a domain of unknown function as Domain Related to Iron, and defined the structure and function of this protein.
Starting from a gene, they’ve discovered an interesting zinc-mirror heme site that plays a key role in sensing iron and regulating metabolism as cyanobacteria photosynthesize. This pins down a piece of the photosynthesis that drives the ocean’s carbon cycles.
Given that similar protein domains exist in plants and algae, these findings could pave the way for insight into other photosynthesizers, as well.
More information:
Nicolas Grosjean et al, A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria, Nature Communications (2024). DOI: 10.1038/s41467-024-47486-z
Provided by
DOE/Joint Genome Institute
Citation:
New insights into how cyanobacterial proteins cycle carbon in changing conditions (2024, June 7)