Belgian researchers from VIB-KULeuven Center for Microbiology and VIB-UGent Center for Plant Systems Biology have developed a new toolbox of 16 different short DNA sequences that allow triggering controlled and specific recombination events in any genome.
This new patented toolbox, complementing—and for certain applications surpassing—CRISPR, is now available for researchers and industry in the field of genome engineering. The results are reported today in two concurrent papers in Nature Communications.
Site-specific recombinases enable efficient cutting and pasting of DNA at specific locations in the genome, where each recombinase recognizes one precise DNA sequence. Due to their sequence-specificity, CRISPR systems have overshadowed site-specific recombinases in the past decade as a genome engineering tool.
CRISPR systems caused a revolution in the field because they can very easily be targeted to different genomic loci. However, because site-specific recombinases operate differently, they circumvent some of the major issues of CRISPR, including the toxicity of DNA double strand breaks causing undesired point mutations and structural variation, the low editing efficiencies faced in many non-conventional organisms and non-dividing cells and the difficulty to insert large DNA fragments. Moreover, the very complex patenting of CRISPR makes its use in research and industry often difficult and expensive.
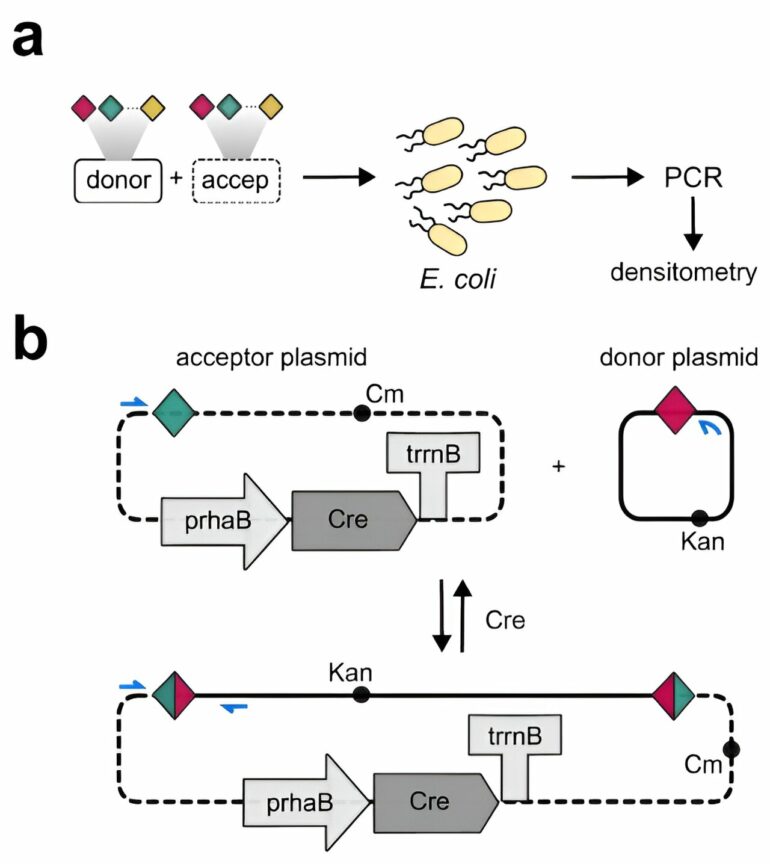
The research groups of VIB-KULeuven Center for Microbiology and their colleagues at VIB-UGent Center for Plant Systems Biology have now addressed the shortcomings of earlier site-specific recombination as a means for genomic engineering. The team has expanded the toolbox that uses a viral recombinase (Cre) so that it can now specifically recognize, cut, and paste multiple DNA sites. The team has identified a set of 16 sites that efficiently recombine with the same site, but not with any other site of this set, and this for different organisms.
“Due to these orthogonal recombination systems, we can avoid the unpredictable way in which multiple recombination sites in a genome interact with each other,” says Kevin Verstrepen, Director of VIB-KULeuven Center for Microbiology. “This opens routes for many research projects to install many small or large genomic edits simultaneously or to repeatedly recycle markers during genome engineering efforts.”
Applications for research and industry
Charlotte Cautereels, Ph.D. student at the lab of Kevin Verstrepen, established the new toolbox in yeast and tested these in bacterial cells. She then collaborated with the VIB-UGent PSB center to also demonstrate its efficiency in plant cells.
In her latest publication, she demonstrates how it can be used to optimize the expression of metabolic pathway genes and production titers of industrially relevant molecules. A single round of shuffling dedicated gene regulators with the new recombination sites already doubled the production titers.
“We were able to corroborate that modifying expression through our recombination-based setup allows for a rapid and efficient gene expression optimization in heterologous biosynthetic pathways,” says Cautereels. “Not only does this new toolbox offer possibilities to improve microbial cell factories, it also validates our toolbox for scientists and industry working in the field of genome engineering.”
More information:
Charlotte Cautereels et al, Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts, Nature Communications (2024). DOI: 10.1038/s41467-024-44996-8
Charlotte Cautereels et al, Combinatorial optimization of gene expression through recombinase-mediated promoter and terminator shuffling in yeast, Nature Communications (2024). DOI: 10.1038/s41467-024-44997-7
Citation:
New toolbox allows engineering of genomes without CRISPR (2024, February 16)