Wnt signaling is a well-known mode of cell-to-cell communication in multicellular biological organisms. It involves the secretion of small Wnt glycoproteins, by signaling cells, that bind to receptor proteins in the membrane of receiving cells. This signal modifies proteins on the inside of these receiving cells to make cells grow, divide or differentiate.
This mode of communication is fundamental in both normal and altered cellular development, such as in cancer and wound healing, and has remained in the limelight for over four decades of research. Some of the open research questions revolve around the extraordinary complexity in the number of Wnt pathway members, functioning both inside, outside, and at the surface of cells, and how different outputs of the pathway are achieved via the use of specific members.
A team of scientists, led by Professor Antónia Monteiro from the Department of Biological Sciences at the National University of Singapore’s (NUS) Faculty of Science, has uncovered some of this complexity by using butterfly wings as a model system. The findings were published in Science Advances on 26 July 2023 with a cover feature.
Butterfly wings function as a large two-dimensional (2D) canvas of cells that “talk” to each other during development to pattern exquisite and detailed color patterns. They are, thus, exceptional systems to explore the role and diversity of Wnt signaling in such patterning mechanisms. This includes exploring where different elements of the signaling pathway are present, what function they are playing in those cells, and how they are likely to interact with each other.
Using state-of-the-art in-situ localisation technologies, the researchers have deciphered the expression patterns of all eight Wnt glycoproteins, and all four membrane receptor proteins (Frizzleds), present in the genome of Bicyclus anynana butterflies.
They have also described the spatial and temporal dynamics of an intra-cellular protein (Armadillo) during wing development and tested the function of this and a few other pathway members using the CRISPR-Cas9 genome editing tool. They showed how different pathway members are likely to spatially and temporally regulate each other to define many of the color patterns in the wings of this butterfly species.
Dynamic role of Wnt signaling in butterfly wing development
Dr. Tirtha Das Banerjee, a Research Fellow at the NUS Department of Biological Sciences, used immunostaining and gene localizing technologies to find that Wnt pathway members are extremely dynamic. Distinct and dynamic bands and circular blotches of multiple proteins from the Wnt pathway were observed as the wing developed. The Armadillo protein, for example, initially present in a homogeneous fashion across all cells of early wings became progressively localized to specific cells including the eyespot centers and wing margin where it plays a role in color differentiation.
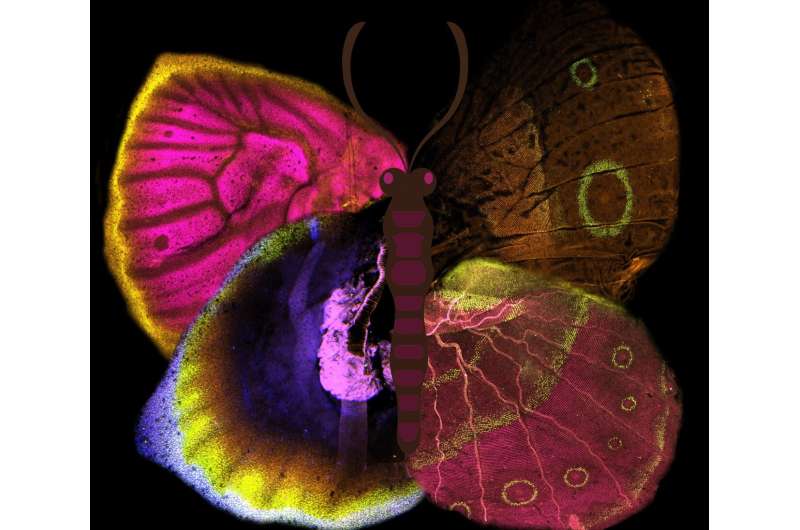
Expression of different Wnt signalling genes, and some of their targets, in the larval (left) and pupal (right) wings of Bicyclus anynana butterflies. © Tirtha Das Banerjee
The gene coding for another Wnt member, WntA, was found to be expressed along a thick band pattern running along the center of the developing wing. When this gene was knocked out by Dr. Heidi Connahs, a Research Fellow at the NUS Department of Biological Sciences, it led to disruptions in both the color and the width of this central band pattern, showing that it functions in these two roles.
Dr. Suriya Murugesan, a Research Fellow at the NUS Department of Biological Sciences, tested the function of Frizzled4 and found that this gene also has a dual role in the differentiation of the eyespot centers and in the orientation of scale cells in the eyespot domain. This family of genes, known as Frizzleds, have a similar role in the orientation of bristles in flies and hairs in mammals.
Interaction between Wnt signaling pathways defines patterns on butterfly wings
One of the key findings of the study was the discovery of the complex spatial and temporal regulation of the different Wnt signaling pathways in patterning the wings of butterflies. Cells expressing distinct members of the Wnt pathways are likely regulating each other and keeping the expression of other members of the pathway shut down to create signal specificity.
Protein frizzled2, for example, is expressed in a very intricate pattern with cells lacking expression of this receptor along the central (wntA), eyespot (frizzled4), and wing margin (frizzled9) domains. These domains, when combined, cover the entire field of cells in the wings of the butterfly.
Prof Monteiro said, “This means that Wnt signaling is taking place everywhere in the wing, but different members of this pathway are controlling different colors and patterns in different areas.”
“We are only beginning to decipher how nature over millions of years of evolution has programmed such complex yet elegant patterning systems using Wnt signaling as a base of our studies,” added Dr. Banerjee.
Dr. Banerjee said, “Many aspects of Wnt signaling in the butterfly system need further exploration, and work in this area is likely to increase our understanding of how this fundamental signaling pathway, used by every multicellular being in our planet, has diversified and evolved into the complex cell-cell communication system we see today. These studies will have profound implications not only in our understanding of color patterns in insect systems but also in other animals where this signaling is conserved.”
More information:
Tirtha Das Banerjee et al, Spatial and temporal regulation of Wnt signaling pathway members in the development of butterfly wing patterns, Science Advances (2023). DOI: 10.1126/sciadv.adg3877
Provided by
National University of Singapore
Citation:
Patchwork of Wnt signaling ligands and receptors pattern the wings of butterflies, researchers discover (2023, August 3)



