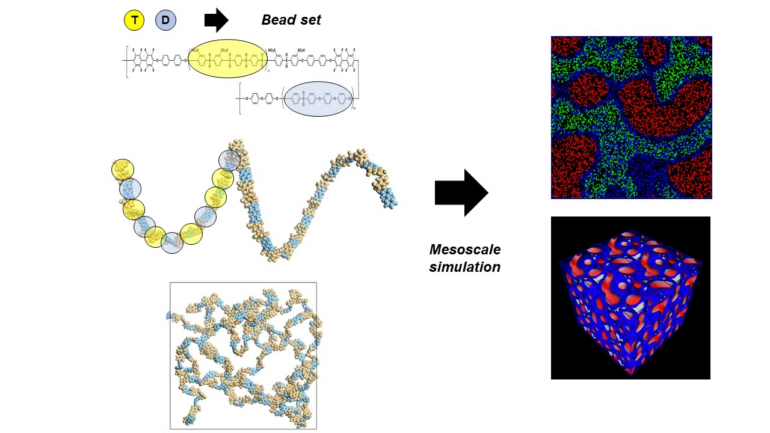
Researchers at the Niels Bohr Institute, University of Copenhagen and University of Southern Denmark have recently published FreeDTS—a shared software package designed to model and study biological membranes at the mesoscale—the scale “in between” the larger macro level and smaller micro level.
This software fills an important missing software among the available biomolecular modeling tools and enables modeling and understanding of many different biological processes involving the cellular membranes, for example, cell division.
Membrane shape contains information about the physiological state of the cell and overall health of an organism, so this new tool, with its wide array of applications, will enhance our understanding of cell behavior and open routes for diagnostics of infections and diseases such as Parkinson’s.
The publication of FreeDTS is now reported in Nature Communications.
Sharing a powerful tool that could have provided NBI with an advantage. Why?
The software package Weria Pezeshkian from the Niels Bohr Institute has been working on for the last five years, after an initial idea between him and John Ipsen from the University of Southern Denmark, is shared—laid open for every researcher in this field to use.
Normally the competition for achieving scientific results is high, and science advancements kept secret until publication—so this seems like a very generous attitude indeed. So generous it might seem a bit naive.
It is a strange mix of respect for the “pioneers” of the biomolecular modeling field and the fact that the field offers so many unanswered questions that it would seem almost disrespectful towards the scientific community to keep the tool to ourselves, Pezeshkian explains.
“There are so many questions and bottlenecks to tackle to reach the end goals, that it would be unlikely that we work on exactly the same problems. However, occasional overlap occurs and is a worthwhile cost we pay for advancing the field.
“But there is another aspect as well: One of the reasons our community, the biomolecular simulation and modeling community has had this surge in popularity and a fast growth is that we’ve always strived to get more people into the game and share ideas, results and methods and often direct assistance without expecting immediate personal gains.
“This culture was built by the early pioneers in the field, for one, late , who always promoted this approach of sharing and bringing people in—so we are to a large extent standing on the shoulders of giants in this respect.”
Biological membranes—what are they really?
When you consider a cell, you can imagine a whole lot of small “factories” inside, called organelles, doing their thing—surrounded by a membrane.
The cell also is surrounded by a membrane called Plasma membrane. But membranes are not just a boundary surface. They are actively participating in many processes. They are made from a myriad of different molecules, and they are dynamic, in motion all the time.
Many diseases are associated with irregular membrane shape and abnormal biomolecular organization, so the study of membranes can help us understand the state of a cell and overall health of an organism. For instance, when a neuron has increased spiking activity, indicating a higher energy demand, the structure of mitochondria, an organelle responsible for generating cellular energy parcels from food (often referred to as the powerhouse of the cell), undergoes changes.
Moreover, certain diseases, e.g., Alzheimer’s for one, have been associated with changes in the mitochondrial membrane shapes.
Computer models will improve our abilities within diagnostics
“For now, we are not able to see exactly what the exact causes of changes in membrane shape are and how are they exactly related to the diagnostics of a certain disease. But at some point, in the future, the try and error works in the lab will become minimal because modeling will guide experiments with unimaginable accuracy, as our modeling becomes more precise and the power of computational options increasing.
“We will need a lot of adjustments and there is still long way to go, so it is really nice to work within this sharing community, because we all work on different aspects of it,” Pezeshkian explains.
“This is probably stretching it a bit far, but possibly, in the future, by imaging for example our mitochondria and leveraging physics-based computer simulations we may be able to say, This person has this disease with this specific genetic deficiency. So, the perspective for computational modeling is rather great—we are not there yet, but we can see it in the horizon.”
More information:
Weria Pezeshkian et al, Mesoscale simulation of biomembranes with FreeDTS, Nature Communications (2024). DOI: 10.1038/s41467-024-44819-w
Provided by
Niels Bohr Institute
Citation:
Physicists develop modeling software to study biological membranes at the mesoscale (2024, March 22)