When it comes to drug development, membrane proteins play a crucial role, with about 50% of drugs targeting these molecules. Understanding the function of these membrane proteins, which connect to the membranes of cells, is important for designing the next line of powerful drugs. To do this, scientists study model proteins, such as bacteriorhodopsin (bR), which, when triggered by light, pump protons across the membrane of cells.
While bR has been studied for half a century, physicists have recently developed techniques to observe its folding mechanisms and energetics in the native environment of the cell’s lipid bilayer membrane.
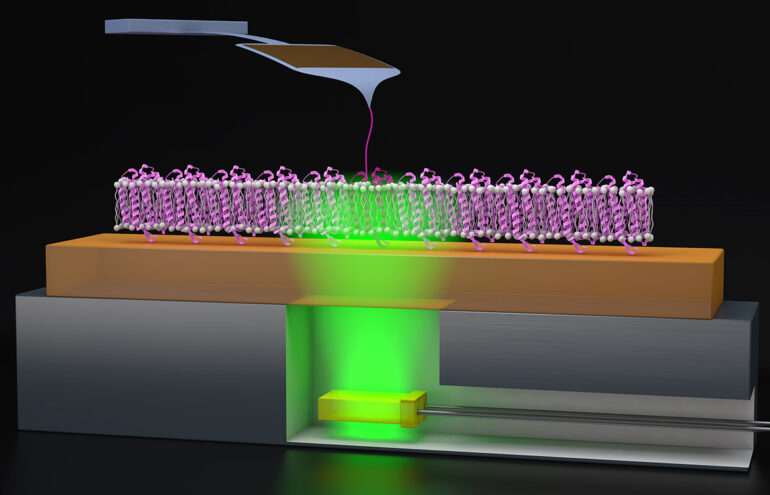
In a new study published by Proceedings of the National Academy of Sciences (PNAS), JILA Fellow Thomas Perkins and his team advanced these methods by combining atomic force microscopy (AFM), a conventional nanoscience measurement tool, with precisely timed light triggers to study the functionality of the protein function in real-time.
“The energetics of membrane proteins has been challenging to study and therefore not well understood,” explained Perkins. “Using AFM and other methods, we can create ways to look into this further.” Armed with a better understanding of the energetics of these proteins, chemists can design drugs that are more potent for specific symptoms and illnesses caused by protein misfunction.
Measuring millisecond protein dynamics
While bR is a microscopic protein, it can be seen by the naked eye, and even in satellite images, when archaeon microorganisms bloom, they leave vast amounts of it as residue in salt-water ponds. “The ponds become filled with what’s called Halobacterium salinarum, the parent organism of bacteriorhodopsin,” Perkins said. “These ponds are used to harvest salt, and because they’re warm and salty, the bacteria love to grow there.”
At the microscopic level, bR works with other membrane proteins to produce energy for the cell by creating a proton gradient on one side of the cell membrane, which ushers the proton through to the other side of the membrane. bR does this by folding and unfolding its helices into specific shapes to control how many protons pass through the membrane. During this process, the proton migration produces chemical energy in the form of adenosine-tri-phosphate (ATP).
For Perkins and his co-author David Jacobson (a former JILA postdoctoral researcher and now an assistant professor at Clemson University), bR presented an opportunity to design a new experimental method for looking at real-time functional energetics.
To study proteins like bR, Jacobson and Perkins utilize AFM, which acts like a tiny finger to pull on the protein gently, which helps the AFM to feel the protein’s surface, mapping out its structure and giving a better understanding of how the protein folds.
Because bR’s folding processes are triggered by light, Perkins and Jacobson added a lighting element to the AFM procedure. “We had this clever idea to glue super thin green LEDs—which trigger the bacteriorhodopsin—to a metal puck, which we can attach to the AFM,” Perkins said. “These green LEDs are also cheap, like $1.00 apiece or $1.50 apiece. Compared to our AFM cantilever, which costs about $80 apiece, throwing away a $1.50 LED is hardly something we worry about.”
With this inexpensive add-on to their AFM, Perkins, and Jacobson could induce the bR to fold and unfold with millisecond precision. After collecting their data, the researchers found that the protein correctly folded 60% of the time, allowing the protons to pass through the membrane.
To verify the energetics and real-time function of the protein folding, the scientists mutated the bR protein to remain always in the “open” or unfolded state. Using their new experimental setup, they could reproduce findings similar to what they observed before in the “open” phase of the bR photocycle.
“In biology, you might see something, but you need to ask, am I seeing what I think I’m seeing?” Perkins said. “So, by making a mutation and seeing the effect that we expected, we have increased confidence that we’re really studying the process we think we are studying.”
The mystery of the misfolded protein
While Perkins and Jacobson observed proper folding 60% of the time, the other 40% of cases surprised them, as the protein misfolded but could still pump a proton through the membrane. “The misfolding is actually stabilizing,” added Perkins. “And that was really surprising.” In many cases, protein misfolding does not result in stabilization.
Due to the energetic stabilization, Perkins and Jacobson theorized that the bR’s structural helices weren’t separating properly to provide a completely open tunnel for the proton, though it still wiggled through, a process difficult to detect with AFM imaging.
Trying to understand the underlying mechanisms for the misfolding better, Perkins and Jacobson lowered the force on the AFM pulling assay to zero to see if this would coax the protein to fold correctly. However, the results remained the same: 40% of cases resulted in misfolding.
These results, with the same amount of misfolding, puzzled the researchers. While Perkins and Jacobson couldn’t identify the cause of these misfolding cases, they hope to investigate further. Now, they are interested in seeing what the rest of the biophysics community makes of these results.
“There could be more subtle effects, or maybe some new science there,” Perkins added. “It could be that there’s a pathway that perhaps people haven’t been able to see before.”
More information:
David R. Jacobson et al, Quantifying a light-induced energetic change in bacteriorhodopsin by force spectroscopy, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2313818121
Citation:
Probing proton pumping: New findings on protein folding in bacteriorhodopsin (2024, February 8)