To discover and thoroughly demonstrate the newly identified noncanonical cleavage mechanism, the Hong Kong University of Science and Technology (HKUST) research team, led by Prof. Tuan Anh Nguyen, Assistant Professor of the Division of Life Science, used several sophisticated techniques, such as miRNA sequencing, pri-miRNA structure analysis, and high-throughput pri-miRNA cleavage assays for approximately 260,000 pri-miRNA sequences.
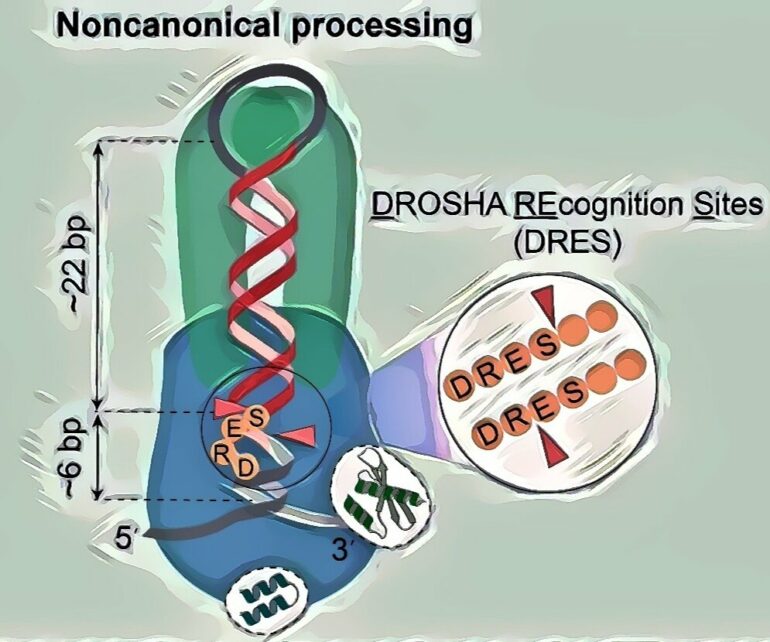
In contrast to the canonical mechanism, the noncanonical mechanism does not rely on several essential protein and RNA elements required for the canonical mechanism. The study also revealed previously unrecognized DROSHA recognition sites (DRES), which are critical for noncanonical cleavage but can also function in the canonical cleavage mechanism.
Furthermore, the study highlights the evolutionary aspect of this noncanonical cleavage mechanism, revealing that it is conserved across several animal species. This finding suggests that the noncanonical mechanism plays a significant role in the evolution of miRNA biogenesis and regulation.
MicroRNAs (miRNAs) are tiny RNA molecules that play a crucial role in regulating the activity of genes. They help control various biological processes, such as cell growth, development, and immunity. In recent years, scientists have been researching extensively on miRNAs to better understand their functions and the mechanisms involved in their production.
Now, scientists at the Hong Kong University of Science and Technology (HKUST) have made a groundbreaking discovery in molecular biology, unveiling a noncanonical cleavage mechanism for the Microprocessor (MP, DROSHA-DGCR8 complex) complex, responsible for producing miRNAs in humans and other animals by processing primary miRNA transcripts (pri-miRNAs).
This groundbreaking discovery sheds light on a long-standing mystery in molecular biology and could have far-reaching implications for our understanding of gene regulation, cellular processes, and the evolution of miRNA biogenesis pathways in animals.
The Microprocessor complex in animals was discovered in 2004, and since then, its molecular mechanisms in producing miRNAs have been extensively studied by many research groups. These studies together built a molecular mechanism model of this enzyme, called the canonical pri-miRNA processing mechanism.
However, this mechanism can only explain how the enzyme cleaves many pri-miRNAs in animals. Since pri-miRNAs in animals are highly diverse in structure and sequences, this mechanism fails to explain how a significant portion of pri-miRNAs are processed.
The noncanonical pri-miRNA processing mechanism, discovered and published in Molecular Cell, solves a two-decade-long mystery in molecular biology regarding the cleavage of numerous pri-miRNAs in animals and complements the previously known canonical mechanism. In simpler terms, this discovery reveals a new way our cells produce miRNAs, which could have implications for our understanding of gene regulation, cellular processes, and the evolution of miRNA biogenesis pathways in animals.
Key findings from HKUST’s study include:
The discovery and comprehensive characterization of a new way cells produce miRNAs in animals, solving a long-standing mystery in molecular biology.The identification of DRES that determine cleavage efficiency and accuracy of MP, opening a new research direction to examine whether other similar enzymes also contain RNA recognition sites.Evidence that the noncanonical miRNA production mechanism is conserved across various animal species, with a significant role in miRNA biogenesis in worms, such as C. elegans and C. briggsae.An explanation for how MP cleaves many short stem pri-miRNAs in animals, suggesting broader cellular functions for the MP complex.
The discovery of the noncanonical cleavage mechanism of the MP complex in miRNA biogenesis has far-reaching implications for future studies in molecular biology. The identification of this mechanism opens up new avenues of research and expands our understanding of the regulatory landscape of miRNA biogenesis in animals.
One of the most significant implications is the potential to discover a greater variety of substrates for the MP complex in animals. Previously, the canonical mechanism was unable to account for the processing of certain pri-miRNAs. Now, with the noncanonical mechanism, researchers can reevaluate RNA substrates that were previously unexplained or overlooked. This could lead to the identification of novel pri-miRNAs and other RNA substrates that are specifically processed by the noncanonical mechanism.
Another implication lies in the potential to reveal new functions of the MP complex in animals. As the noncanonical mechanism can process short stem pri-miRNAs, it suggests a broader cellular role for the MP complex. This could lead to the discovery of previously unknown roles in gene regulation and cellular processes, such as development, differentiation, and immunity.
Finally, by demonstrating the conservation of the noncanonical mechanism across various animal species, particularly in worms like C. elegans and C. briggsae, this study highlights the evolutionary significance of this mechanism. Future studies could delve deeper into the evolutionary aspects of both canonical and noncanonical mechanisms, shedding light on the development and diversification of miRNA biogenesis pathways in animals.
In conclusion, the discovery of the noncanonical mechanism in miRNA biogenesis has opened up new research possibilities and expanded our understanding of the molecular mechanisms underlying miRNA production. This paves the way for future discoveries in molecular biology, leading to a better understanding of gene regulation, cellular processes, and the evolution of miRNA biogenesis pathways in animals.
More information:
Thuy Linh Nguyen et al, Noncanonical processing by animal Microprocessor, Molecular Cell (2023). DOI: 10.1016/j.molcel.2023.05.004
Provided by
Hong Kong University of Science and Technology
Citation:
Researchers unveil long-sought noncanonical cleavage mechanism in miRNA biogenesis (2023, June 2)