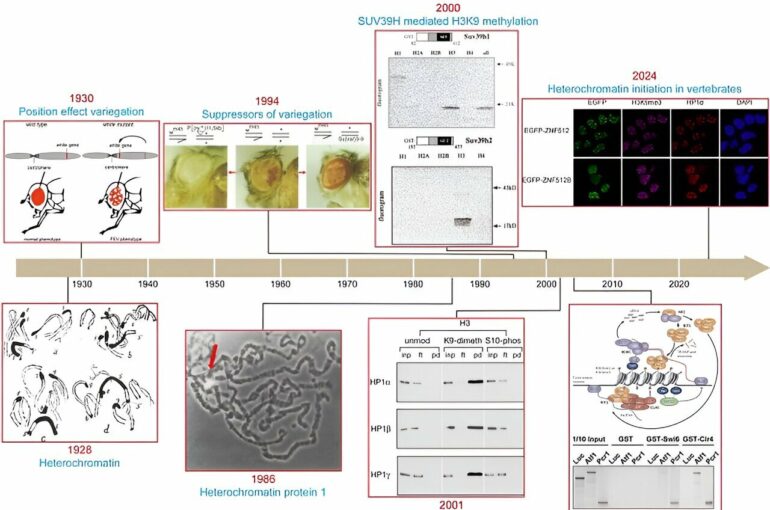
A study published in Nature on July 4 by Prof. Zhu Bing from the Institute of Biophysics of the Chinese Academy of Sciences has shed light on the conserved mechanism responsible for the initiation of pericentric heterochromatin in vertebrates.
In vertebrates, the mechanism of pericentromeric heterochromatin assembly remains poorly understood. To identify factors responsible for the establishment of pericentric heterochromatin that may have been overlooked in previous studies based on genetic screening, the researchers developed a technique capable of identifying the proteome near specific genomic loci.
This technique is based on the CRISPR/Cas9 system with specific targeting capabilities and the APEX2 system with proximity labeling capabilities. Using this technology, they were able to identify the proteome near pericentric heterochromatin in mouse embryonic stem cells.
The researchers found that the zinc finger proteins ZNF512 and ZNF512B are localized at the pericentric heterochromatin regions by their specific binding to pericentric heterochromatin DNA.
Furthermore, whether in exogenously introduced repetitive regions or in pericentric heterochromatin regions, zinc finger proteins ZNF512 and ZNF512B can recruit them to specific sites through direct interaction with SUV39H1 and SUV39H2, resulting in histone H3K9me3-mediated heterochromatin.
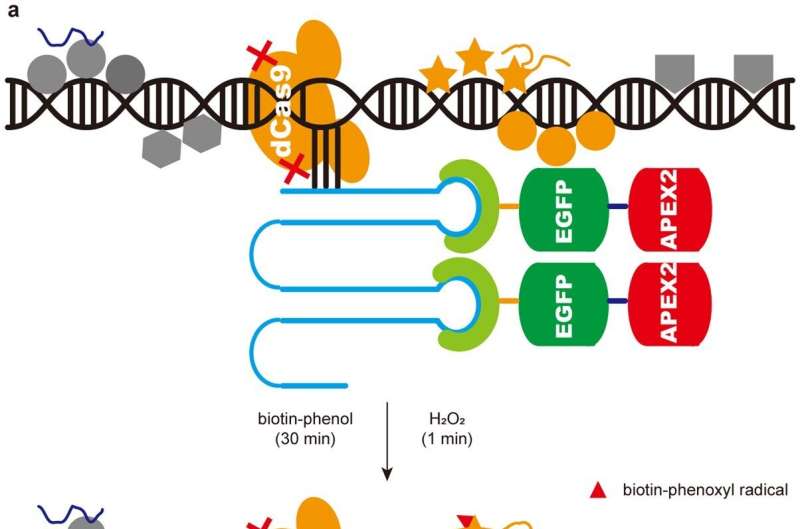
Workflow and the overlap analysis to characterize the protein composition of pericentric heterochromatin. © Nature (2024). DOI: 10.1038/s41586-024-07640-5
They demonstrated that ZNF512 and ZNF512B from different species can specifically target pericentric heterochromatin regions in other vertebrates. This ability is attributed to the identical zinc finger motifs and conserved longer linkers between the zinc fingers of ZNF512 and ZNF512B. The same zinc finger motifs provide the ability to recognize repetitive DNA, while the longer linkers provide the flexibility to recognize non-contiguous DNA sequences.
This study reveals the de novo establishment mechanism of constitutive pericentric heterochromatin and explains why apparently non-conserved pericentric heterochromatin sequences in vertebrates still have the same histone H3K9me3 modification.
In addition, the discovery that zinc finger proteins with split zinc fingers can recognize non-contiguous DNA sequences unveils a novel DNA recognition pattern, which has important implications for the study of protein-DNA recognition patterns.
The results may lead to the discovery of more proteins with such characteristics and inspire bioinformaticians to develop new algorithms for calculating DNA binding motifs.
More information:
Runze Ma et al, Targeting pericentric non-consecutive motifs for heterochromatin initiation, Nature (2024). DOI: 10.1038/s41586-024-07640-5
Provided by
Chinese Academy of Sciences
Citation:
Scientists uncover conserved mechanism of pericentric heterochromatin initiation in vertebrates (2024, July 5)