The cost of sequencing has gone down tremendously. But still it is not used in daily practice. One of the reasons for this is that the processing of the raw data into useful insights takes a long time (several days for whole genome sequencing) and requires a lot of expensive resources (such as servers that need to be rented in a data center).
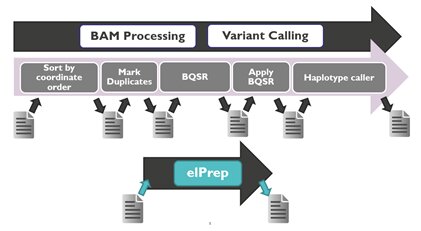
“The problem with current software for sequencing data and variant calling is that it is not structured in the best way,” explains Charlotte Herzeel from imec’s ExaScience Life Lab. “Every step in the pipeline (e.g. mark duplicates, base quality score recalibration or BQSR) is done by separate software tools that each require in- and output to a memory source and can only be executed when the former step is finalized. With our new software platform, we rethought this process and considered the pipeline as a whole.”
elPrep5: Parallelization for more efficient variant calling
ElPrep5 is the final version of the software platform that imec’s ExaScience Life Lab developed for the sequencing pipeline. This final update also includes the variant calling step, a step that typically takes up a substantial part of the total runtime (38–80%). ElPrep is developed in Linux and written in Go (a programming language developed by Google). It is released both as an open-source project on GitHub and as a premium license with support.
“The software optimization strategy consists of merging the execution of multiple pipeline steps, parallelizing their execution, and avoiding file I/O,” explains Herzeel. “It produces results like established state-of-the-art genome analysis programs such as SAMtools, Picard and GATK4. This is important from a user’s perspective as it allows elPrep5 to be used as a drop-in replacement for other popular tools.”
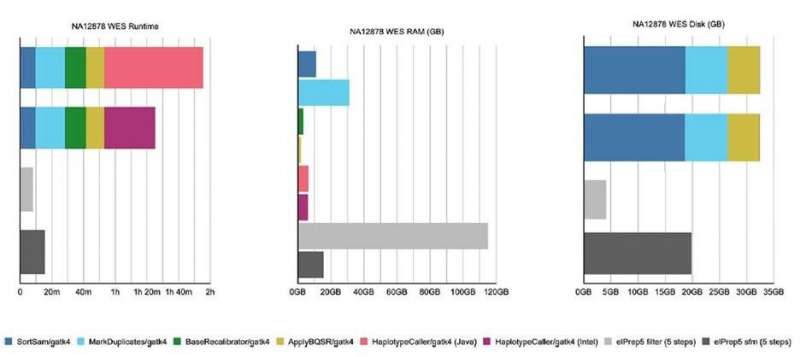
Figure 2. Results from the benchmarking experiment on whole exome data, using both GATK4 and elPrep software. © IMEC
Shifting up a gear: 8 to 16 times faster results
Several experiments were done to benchmark the efficiency of the elPrep5 software. “In a first experiment, a whole exome sequencing was performed using a 96CPUx384GB server,” says Herzeel. “The data were either sequenced with the widely-used GATK4 or the elPrep software. For GATK4, two modes of the software were used: the Java (the standard haplotype caller algorithm) and Intel (algorithm optimized for parallelization) mode. Also, for elPrep, two modes were tested: the filter (loading all input data into RAM to avoid intermediate I/O to disk) and sfm (splits up the data by chromosomal regions) mode. Overall, the filter mode is useful for smaller data sets but uses more RAM. Whether the filter mode can be used depends on the size of the input BAM in relation to the available RAM.”
The results of this experiment are shown in figure 2 and 3.
On the whole exome data, the elPrep filter mode:
is 11 to 15 times faster than GATK4 (Java and Intel mode)uses 4 times more RAM than GATK4 (Java and Intel mode)uses almost 10 times less disk space than GATK4 (Java and Intel mode)
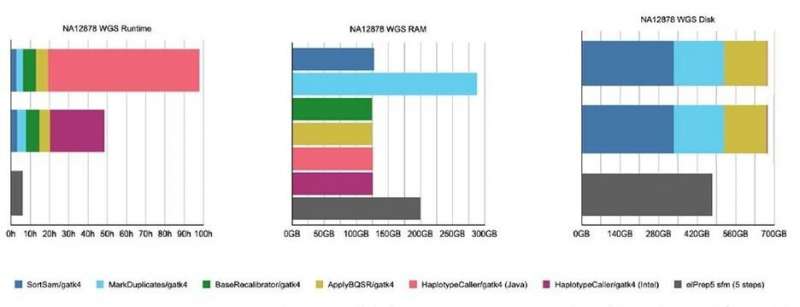
Figure 3. Results from the benchmarking experiment on whole genome data, using both GATK4 and elPrep software. © IMEC
On the exome data, the elPrep sfm mode:
is 6 to 7.5 times faster than GATK4 (Java and Intel mode)uses half the RAM of GATK4 (Java and Intel mode)uses half the disk of GATK4 (Java and Intel mode)
“In the context of a cloud setup, the filter mode is overall the cheapest and most efficient mode to process the data, because, even though it uses more RAM than the sfm mode, the runtime is reduced so much that it reduces the server rental cost,” explains Herzeel.
For the whole genome data, a similar test was done. The only difference was that for elPrep only the sfm mode was used since the filter mode is not suitable for such large data sets.
On the genome data, the elPrep sfm mode:
is 16 times faster than GATK4 Java mode and 8.5 times faster than the Intel modeuses 70% RAM of GATK4 (Java and Intel mode)uses 70% of the disk of GATK4 (Java and Intel mode)
“ElPrep 5 shows more speedup for whole-genome data (16x) than for whole-exome data,” summarizes Herzeel. “This is because variant calling, which is included in elPrep 5, typically takes up a larger portion of the overall runtime of a pipeline for whole genome data. Hence more time is spent proportionally on variant calling for whole-genome data and there is more computation for elPrep to speed up.”
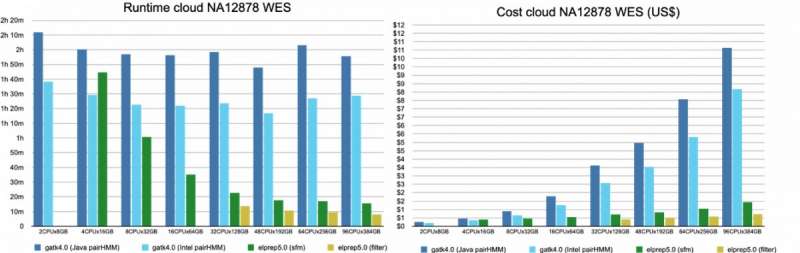
Figure 4. The runtime and cost for running the variant calling pipeline on whole-exome data, on a variety of servers. © IMEC
The more resources, the faster you get (with elPrep)
Another experiment was done to study the ‘scaling’ potential of the elPrep software: how well does it perform in terms of reducing runtime when more hardware resources can be used. “If software scales well, the higher cost of renting servers with more computational resources can be compensated by the reduction in runtime,” comments Herzeel. “To do this test, the pipeline was executed on different servers with varying numbers of CPUs and RAM, using the widely-used GATK4 as reference.”
Results are shown in figure 4 (whole-exome) and figure 5 (whole-genome).
For whole-exome data, elPrep (filter and sfm mode) scales very well. The runtime nearly halves for each increase of resources. Some concrete results that can be derived from the graphs:
the fastest elPrep run is a filter mode on 96CPUx384GB. This is some ten times faster and 5 times cheaper than the fastest run with GATK4, Intel mode on 48CPUx192GBthe cheapest run is with GATK4, Intel mode on 2CPUx8GB. However, it is more than 12 times slower than the fastest elPrep run while 4 times cheaper. For two times the price of the cheapest GATK4 run, you get a run with elPrep, sfm mode on 8CPUx32GB, that is two times faster.If the user prefers the output of the GATK4 Java mode, then it is cheaper and faster to use elPrep: the run on 32CPUx128GB with elPrep filter mode is slightly cheaper and almost nine times faster than the GATK4 Java mode run on 2CPUx8GB.
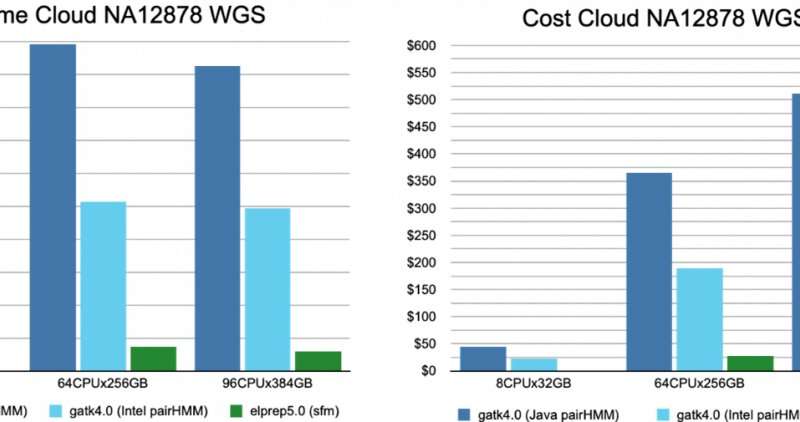
Figure 5. The runtime and cost for running the variant calling pipeline on whole-genome data, on a variety of suitable servers. © IMEC
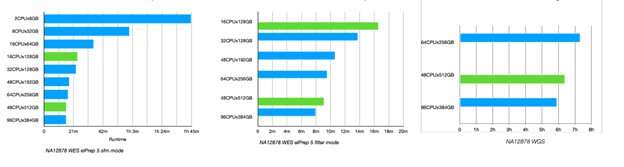
Figure 6. Results of an experiment with elPrep running on both cloud (blue) and Pure Storage (green) infrastructure (using Intel and AMD-based servers). The servers are ranked on amount of RAM and CPUs. With the cloud provider the data resides on their default ‘hot’ data storage, whereas with Pure Storage, the data resides on a Flashblade server connected to the compute servers. The two graphs on the left represent analysis of whole-exome data while the outer right is done on whole-genome data. © IMEC
For whole-genome data, only the sfm mode of elPrep was used, and the software was only run on servers with enough RAM and disk space for this large amount of data. One interesting lesson from the graphs in figure 4 is that the elPrep run on 96CPUx384GB is cheaper than the GATK4 Java run on 8CPUx32GB because the elPrep run is almost 14 times faster. More specifically, the cost goes down from 45 to 32 dollar and the runtime from almost 80h to less than 6h.
Next to servers from a large cloud provider, a test was also done on a system using Pure Storage infrastructure with a different storage architecture (FlashBlade). Results are shown in Figure 6.
Herzeel: “The experiment shows that the elPrep experiments on the Pure Storage infrastructure scale similarly to the cloud benchmarks. This suggests that the Flashblade storage solution of Pure Storage performs at least equally well to cloud provider’s storage solutions.”
ElPrep, and its latest update elPrep5 in particular, is a software for the analysis of sequencing data, including variant calling. It can be used as replacement for established state-of-the-art genome analysis programs such as a.o. SAMtools, Picard and GATK4.
“Our benchmark experiment shows that elPrep 5 speeds up the pipeline execution by a factor 8 to 16x as compared to GATK4,” concludes Herzeel. “Concretely, elPrep 5 executes the variant calling pipeline in less than 6h for a whole-genome sample, and needs less than 8 minutes for a whole-exome sample. ElPrep achieves these speedups using algorithmic innovations and parallelization, runs on regular CPU-based servers without specialized accelerators, and uses fewer RAM and disk resources.”
More information:
Charlotte Herzeel et al, Multithreaded variant calling in elPrep 5, PLOS ONE (2021). DOI: 10.1371/journal.pone.0244471
Citation:
Software optimizations make variant calling 8 to 16 times faster for genome sequencing (2021, December 3)