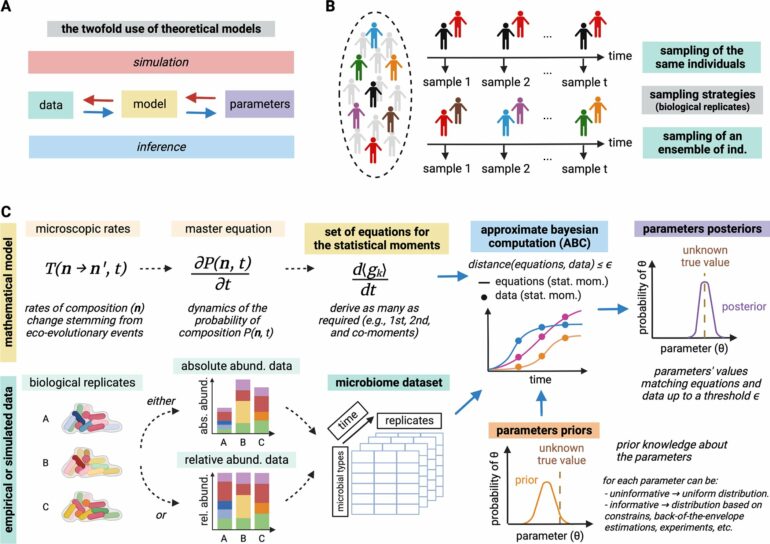
Traditional models for analyzing microbial communities often rely on average microbial abundance over time. While these models can predict temporal changes effectively, they often fail to capture the actual interactions between microbes.
An international research team, including scientists from the Max Planck Institute for Evolutionary Biology (MPI-EB), University College London (UCL), and the University of Aix-Marseille, all involved in the SFB “Metaorganisms” at CAU Kiel, has now addressed these challenges and developed a method that goes far beyond this traditional approach.
“Our model not only takes averages into account but also incorporates variability and correlations between data points, enabling us to capture microbial interactions much more precisely,” explains Dr. Román Zapién-Campos, PostDoc at UCL and lead author of the study published in PLOS Biology.
The researchers developed a stochastic model based on microscopic transition rates—such as birth, migration, or mutations—and calculated the statistical moments of the microbiome composition. This approach allows both the parameters and their uncertainties to be determined. It not only provides more reliable predictions about the dynamics of microbial communities but also enables the detailed identification of specific interactions between microbes.
“Our method bridges a critical gap between metagenomic data and ecological models,” says co-author Arne Traulsen, Director of the Department of Theoretical Biology at MPI-EB. Notably, the approach works with both relative abundance data—typically found in metagenomic studies—and absolute abundance data. This significantly broadens its applicability and allows for analyses regardless of the type of data available.
The approach was successfully applied to simulated data, a key test for the reliability of the method. Additionally, a simplified mouse microbiome, consisting of twelve well-characterized species, was analyzed.
“We were able to uncover not only the underlying mechanisms of these microbial communities but also to precisely quantify the uncertainties in the model parameters,” explains co-author Dr. Florence Bansept, Group Leader and CNRS Researcher at the University of Aix-Marseille.
This study represents a significant advancement in the understanding of microbial ecosystems and their interactions with their hosts. The newly developed method offers promising prospects for better understanding microbiomes in medicine, as well as microbial communities in the environment.
More information:
Román Zapién-Campos et al, Stochastic models allow improved inference of microbiome interactions from time series data, PLOS Biology (2024). DOI: 10.1371/journal.pbio.3002913
Provided by
Max Planck Society
Citation:
Stochastic model captures microbiome interactions more precisely (2024, December 5)