A team led by UT Southwestern Medical Center researchers has discovered a new way that cells regulate senescence, an irreversible end to cell division. The findings, published in Cell, could one day lead to new interventions for a variety of conditions associated with aging, including neurodegenerative and cardiovascular diseases, diabetes, and cancer, as well as new therapies for a collection of diseases known as ribosomopathies.
“There is great interest in reducing senescence to slow or reverse aging or aging-associated diseases. We discovered a noncoding RNA that when inhibited strongly impairs senescence, suggesting that it could be a therapeutic target for conditions associated with aging,” said Joshua Mendell, M.D., Ph.D., Professor of Molecular Biology and a member of the Harold C. Simmons Comprehensive Cancer Center at UT Southwestern. He is also a Howard Hughes Medical Institute Investigator.
Dr. Mendell led the study with co-first authors Yujing Cheng, Ph.D., a recent graduate of the Genetics, Development, and Disease graduate program; and Siwen Wang, M.D., a former postdoctoral researcher, both in the Mendell Lab.
Cellular senescence is a “double-edged sword,” Dr. Mendell explained. Cells sometimes undergo senescence when a cancer-causing mutation arises, halting uncontrolled cell division and preventing tumors from developing. On the other hand, too much senescence contributes to aging and degenerative diseases.
The Mendell Lab has long studied noncoding RNAs, finding new roles for these molecules in both health and disease. In this newest study, he and his colleagues used a technique for regulating gene activity called CRISPR interference to individually inactivate thousands of noncoding RNAs in human cells that carried a cancer-causing mutation. Usually, this mutation prompts cells to become senescent; however, inactivating a noncoding RNA involved in senescence caused the cells to continue dividing.
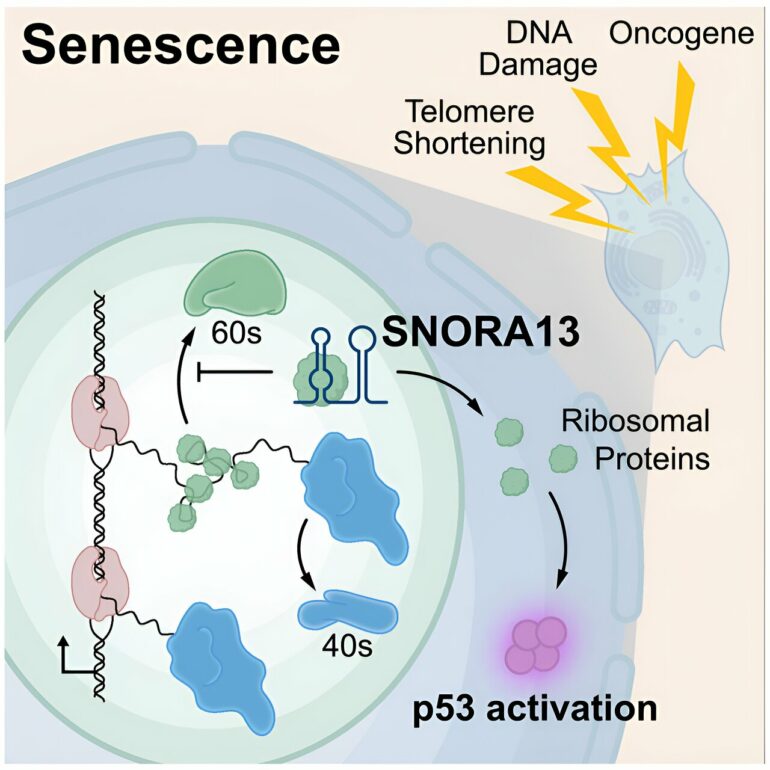
These experiments quickly revealed a previously unrecognized regulator of senescence called SNORA13, a member of a family of noncoding RNAs known as small nucleolar RNAs that are thought largely to function as guides for chemical modification of other RNA molecules. A series of additional experiments showed that SNORA13 plays another important and unexpected role: slowing down the construction of ribosomes, cellular machines that synthesize proteins.
Dr. Mendell explained that cellular stress—prompted by a cancer-causing mutation, for example—can perturb ribosome assembly and push cells into senescence. However, removing SNORA13 caused cells to ramp up ribosome assembly, blocking the quality control that would normally trigger senescence and allowing cells to continue dividing.
Learning more about this process could eventually help researchers control it, Dr. Mendell said. For example, developing drugs that push cells into senescence could offer a new way to treat cancer. Conversely, developing drugs that prevent senescence could slow aging and diseases that typically accompany it, such as cardiovascular diseases, neurodegenerative diseases, and diabetes.
In addition, because of SNORA13’s essential function in regulating ribosome assembly, targeting this noncoding RNA could someday be used to treat ribosomopathies, diseases characterized by abnormal ribosome production or function, such as Treacher Collins syndrome or Diamond-Blackfan anemia.
More information:
Yujing Cheng et al, A non-canonical role for a small nucleolar RNA in ribosome biogenesis and senescence, Cell (2024). DOI: 10.1016/j.cell.2024.06.019
Provided by
UT Southwestern Medical Center
Citation:
Study identifies RNA molecule that regulates cellular aging (2024, July 17)