RNA, specifically mRNA, has enjoyed a bit of publicity recently as the main component of the COVID-19 vaccines.
Its typical job in an organism is to take the genetic material coded in the DNA inside a cell’s nucleus into the interior of the cell where it is transformed into proteins that carry out different functions in the body. Before this happens, however, the RNA is modified to make it more stable and mature.
One of the most common and important RNA modifications in animal cells is called N6-methyladenosine, or m6A. Scientists over the past decade have revealed that the m6A modification has a wide variety of functions, from the programming of cells to the development of embryos.
A major goal for researchers in the field of computational medicine has been to describe the transcriptome—the sum of all of the RNA readouts of a cell—including modifications like m6A. Having this readout can tell scientists when genes are turned off and on in a cell, deciding in large part the cell’s ultimate fate.
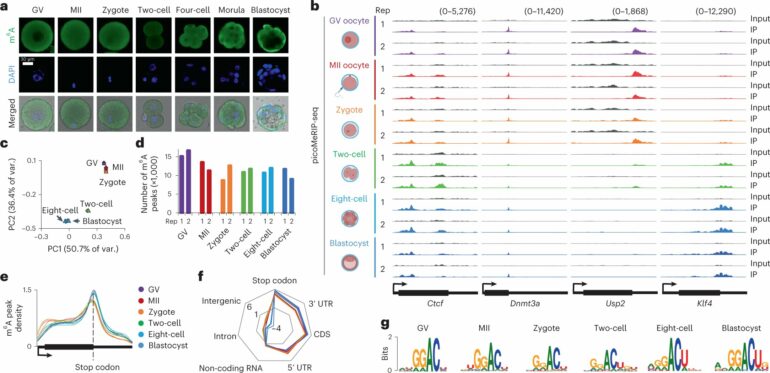
Research from University of Michigan Medical School’s Kin Fai Au, Ph.D. of the Department of Computational Medicine and Bioinformatics, has successfully constructed the landscape of m6A in a mouse embryo, one of the last frontiers for mapping m6A modification.
“Because of the limited amount of cellular material in early embryonic development, it is very challenging to map m6A modification,” said Au. For example, the amount of RNA being studied is measured in picograms, one-trillionth of a gram.
Together with his collaborators Arne Klungland, Ph.D., and John Arne Dahl, Ph.D., of Oslo University in Norway, his team accomplished their study by pooling together mouse embryos at six developmental stages, generating transcriptome-wide m6A maps for each.
Doing so, they discovered intriguing patterns of m6A modification on genes inherited from the mother versus those activated from the developing embryo. They also found m6A distribution in so called transposable elements, previously thought to be ‘junk’ because they do not encode proteins, in the developing embryo.
This study fills a gap in the transcriptome field and sets up researchers to look further into the function of m6A during development, said Au. “Now we have more comprehensive understanding of the different layers of -omics in early embryo development.”
The paper is published in the journal Nature Structural & Molecular Biology.
More information:
Yunhao Wang et al, The RNA m6A landscape of mouse oocytes and preimplantation embryos, Nature Structural & Molecular Biology (2023). DOI: 10.1038/s41594-023-00969-x
Provided by
University of Michigan
Citation:
Study reveals pivotal RNA modification in mouse embryos (2023, April 21)