Context matters. It’s true for many facets of life, including the tiny molecular machines that perform vital functions inside our cells.
Scientists often purify cellular components, such as proteins or organelles, in order to examine them individually. However, a new study published today in the journal Nature suggests that this practice can drastically alter the components in question.
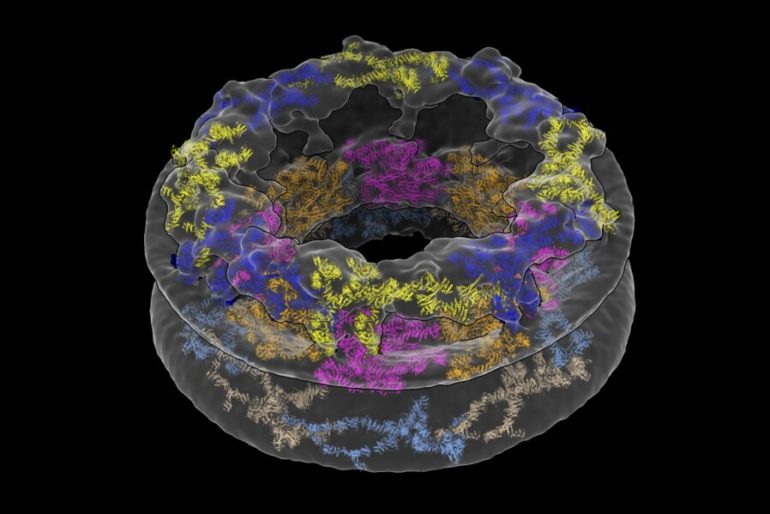
The researchers devised a method to study a large, donut-shaped structure called the nuclear pore complex (NPC) directly inside cells. Their results revealed that the pore had larger dimensions than previously thought, emphasizing the importance of analyzing complex molecules in their native environments.
“We’ve shown that the cellular environment has a significant impact on large structures like the NPC, which was something we weren’t expecting when we started,” says Thomas Schwartz, the Boris Magasanik Professor of Biology at MIT and the study’s co-senior author. “Scientists have generally thought that large molecules are stable enough to maintain their fundamental properties both inside and outside a cell, but our findings turn that assumption on its head.”
In eukaryotes like humans and animals, most of a cell’s DNA is stored in a rounded structure called the nucleus. This organelle is shielded by the nuclear envelope, a protective barrier that separates the genetic material in the nucleus from the thick fluid filling the rest of the cell. But molecules still need a way to come in and out of the nucleus in order to facilitate important processes, including gene expression. That’s where the NPC comes in. Hundreds—sometimes thousands—of these pores are embedded in the nuclear envelope, creating gateways that allow certain molecules to pass.
The study’s first author, former MIT postdoc Anthony Schuller, compares NPCs to gates at a sports stadium. “If you want to access the game inside, you have to show your ticket and go through one of these gates,” he explains.
The NPC may be tiny by human standards, but it’s one of the largest structures in the cell. It’s comprised of roughly 500 proteins, which has made its structure challenging to parse. Traditionally, scientists have broken it up into individual components to study it piecemeal using a method called X-ray crystallography. According to Schwartz, the technology required to analyze the NPC in a more natural environment has only recently become available.
Together with researchers from the University of Zurich, Schuller and Schwartz employed two cutting-edge approaches to solve the pore’s structure: Cryo-focused ion beam (cryo-FIB) milling and cryo-electron tomography (cryo-ET).
An entire cell is too thick to look at under an electron microscope. But the researchers sliced frozen colon cells into thin layers using the cryo-FIB equipment housed at MIT.nano’s Center for Automated Cryogenic Electron Microscopy and the Koch Institute for Integrative Cancer Research’s Peterson (1957) Nanotechnology Materials Core Facility. In doing so, the team captured cross-sections of the cells that included NPCs, rather than simply looking at the NPCs in isolation.
“The amazing thing about this approach is that we’ve barely manipulated the cell at all,” Schwartz says. “We haven’t perturbed the cell’s internal structure. That’s the revolution.”
What the researchers saw when they looked at their microscopy images was quite different from existing descriptions of the NPC. They were surprised to find that the innermost ring structure, which forms the pore’s central channel, is much wider than previously thought. When it’s left in its natural environment, the pore opens up to 57 nanometers—resulting in a 75 percent increase in volume compared to previous estimates. The team was also able to take a closer look at how the NPC’s various components work together to define the pore’s dimensions and overall architecture.
“We’ve shown that the cellular environment impacts NPC structure, but now we have to figure out how and why,” Schuller says. Not all proteins can be purified, he adds, so the combination of cryo-ET and cryo-FIB will also be useful for examining a variety of other cellular components. “This dual approach unlocks everything.”
“The paper nicely illustrates how technical advances, in this case cryo-electron tomography on cryo-focused ion beam milled human cells, provide a fresh picture of cellular structures,” says Wolfram Antonin, a professor of biochemistry at RWTH Aachen University in Germany who was not involved in the study. The fact that the diameter of the NPC’s central transport channel is larger than previously thought hints that the pore could have impressive structural flexibility. “That may be important for the cell to adapt to increased transport demands,” Antonin explains.
Next, Schuller and Schwartz hope to understand how the size of the pore affects which molecules can pass through. For instance, scientists only recently determined that the pore was big enough to allow intact viruses like HIV into the nucleus. The same principle applies to medical treatments: Only appropriately-sized drugs with specific properties will be able to access the cell’s DNA.
Schwartz is especially curious to know whether all NPCs are created equal, or if their structure differs between species or cell types.
“We’ve always manipulated cells and taken the individual components out of their native context,” he says. “Now we know this method may have much bigger consequences than we thought.”
Scientists capture the moving parts of the portal to the cell’s nucleus
More information:
Thomas Schwartz, The cellular environment shapes the nuclear pore complex architecture, Nature (2021). DOI: 10.1038/s41586-021-03985-3. www.nature.com/articles/s41586-021-03985-3
Provided by
Massachusetts Institute of Technology
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.
Citation:
Cellular environments shape molecular architecture (2021, October 13)
retrieved 14 October 2021
from https://phys.org/news/2021-10-cellular-environments-molecular-architecture.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.