A “window to evolution” has opened after mathematicians uncovered the universal explanatory framework for how molecules interact with one another to adapt to new and variable conditions while maintaining tight control over key survival properties.
Landmark research published in Nature Communications by mathematicians Dr. Robyn Araujo at QUT and Professor Lance Liotta of George Mason University in the U.S. sets out the definitive picture of biological adaptation at the level of intermolecular interactions.
Dr. Araujo, from the QUT School of Mathematical Sciences, said the research findings represented a blueprint for adaptation-capable signaling networks across all domains of life and for the design of synthetic biosystems.
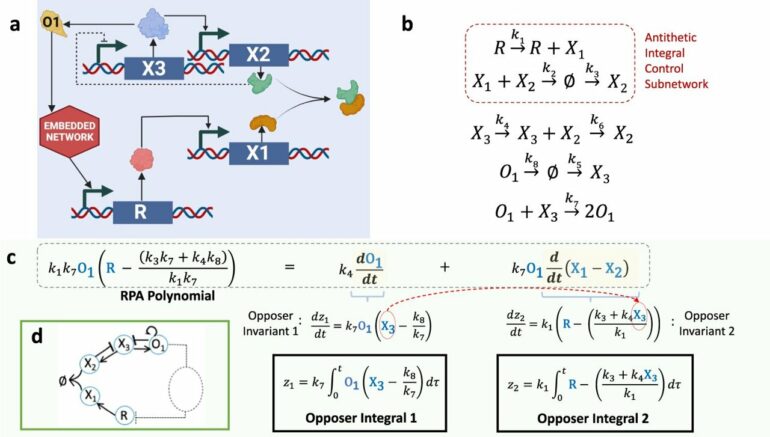
“Our study considers a process called robust perfect adaptation (RPA) whereby biological systems, from individual cells to entire organisms, maintain important molecules within narrow concentration ranges despite continually being bombarded with disturbances to the system,” Dr. Araujo said.
“Until now, no one had a general way to explain how this vital process was orchestrated at the molecular level through the vast, complex, often highly intricate networks of chemical reactions among different types of molecules, mostly proteins.
“We have now solved this problem, having discovered fundamental molecular-level design principles that organize all forms of biological complexity into robustness-promoting, and ultimately, survival-promoting, chemical reaction structures.”
Dr. Araujo said they had found that collections of interacting molecules in living systems cannot simply “transmit” biochemical signals but must actually make “computations” on these signals.
“These complex intermolecular interactions must implement a special type of regulation known as integral control—a design strategy known to engineers for almost a century.
“However, signaling networks in nature are vastly different, having evolved to rely on the physical interactions between discrete molecules. So, nature’s ‘solutions’ operate through remarkable and highly intricate collections of interactions, without engineering’s specially designed, integral-computing components, and often without feedback loops.
“We show that molecular network structures use a form of integral control in which multiple independent integrals, each with a very special and simple structure, can collaborate to confer the capacity for adaptation on specific molecules.
“Using an algebraic algorithm based on this finding, we have been able to demonstrate the existence of embedded integrals in biologically important chemical reaction networks whose ability to exhibit adaptation could never before be explained by any systematic method.”
Professor Liotta said the quest to uncover the fundamental design principles of biological systems throughout nature is considered to be one of the most important and far-reaching grand challenges in the life sciences.
“On the basis of this ground-breaking new research, RPA currently stands alone as a keystone biological response for which there now exists a universal explanatory framework.
“It’s a framework that imposes strict and inviolable design criteria on arbitrarily large and complex networks, and one that now accounts for the subtleties of intricate intermolecular interactions at the network microscale.
“At a practical level, this discovery could provide a completely fresh approach to tackle grand challenges in personalized medicine such as cancer drug resistance, addiction, and autoimmune diseases.”
More information:
Robyn P. Araujo et al, Universal structures for adaptation in biochemical reaction networks, Nature Communications (2023). DOI: 10.1038/s41467-023-38011-9
Provided by
Queensland University of Technology
Citation:
Math unlocks molecular interactions that open window to how life evolved (2023, April 27)