Researchers have developed a new algorithm for recovering the 3D refractive index distribution of biological samples that exhibit multiple types of light scattering. The algorithm helps optimize a new imaging approach called intensity diffraction tomography (IDT).
Jiabei Zhu from Boston University will present this research at the Optica Imaging Congress. The hybrid meeting will take place 14—17 August 2023 in Boston, Massachusetts.
“3D quantitative phase imaging (QPI) has superior features for various applications in the field of biomedical imaging. As a label-free technique, QPI can image transparent living organisms and cells without exogenous contrast agents and dyes which induce phototoxic effects damaging the sample,” explains Zhu.
“Compared with traditional phase-contrast and differential interference contrast microscopy, QPI not only provides high-contrast morphological information but gives quantitative phase information as well. Specifically, 3D QPI can provide high-resolution 3D refractive index (RI) distribution inside the samples. This valuable information can facilitate the research on hematology, neurology, and immunology, helping the diagnosis of disease and infection.”
Although 3D imaging techniques can be used to study thick biological samples, achieving both high-speed acquisition and high resolution is challenging. IDT approaches are label-free phase tomography techniques that help overcome this limitation. They can be performed using a programmable LED array that is easily added to a standard microscope.
Zhu’s research team recently developed two IDT methods known as annular IDT (aIDT) and multiplexed IDT (mIDT) that boost the image acquisition speed enough to visualize dynamic biological samples. Annular IDT (aIDT) uses an LED ring that matches the objective’s numerical aperture, and multiplexed IDT (mIDT) uses multiple LEDs to illuminate the sample simultaneously.
When the researchers discovered that existing IDT reconstruction algorithms did not work well with their new approaches due to the use of high-NA objectives, they decided to develop a new algorithm. It uses a multiple scattering model based on the split-step non-paraxial (SSNP) method, which was recently developed to overcome similar limitations in optical diffraction tomography.
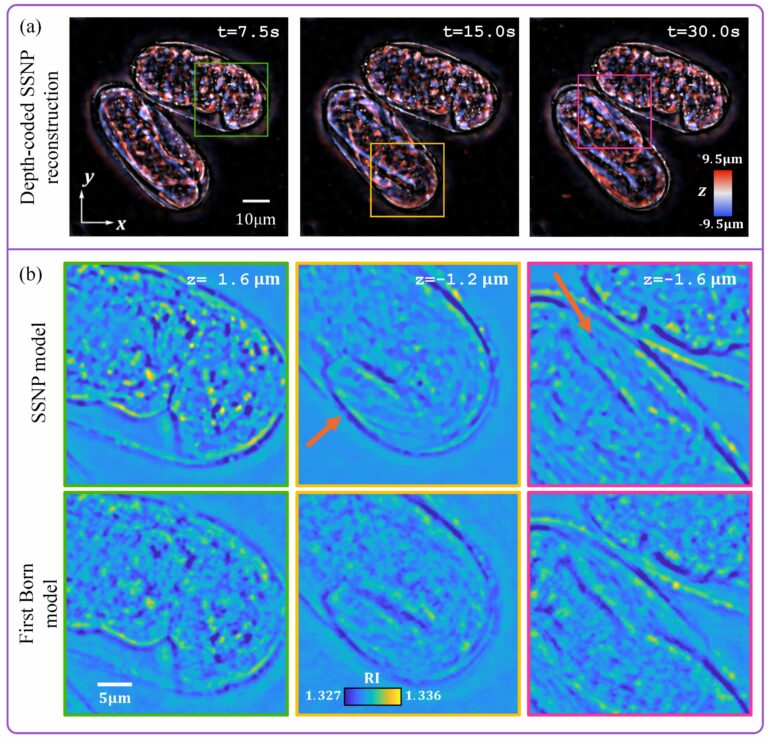
The researchers showed that applying the new IDT reconstruction algorithm to buccal epithelial cells using aIDT allowed easy discrimination of cells at different depths, reconstruction of the cell boundaries and membrane, and visualization of native bacteria around the cells.
They also applied it to a thick multi-scattering live C. elegans embryo using mIDT. The resulting reconstructed images showed details of how the worms were folded, and the single-depth cross-section showed the morphological details of the cells’ outline, the buccal cavity and the tail of the worm.
Overall, the experiments showed that by extending the SSNP method to IDT, the researchers were able to achieve high-quality images with a large field of view.
Citation:
New algorithm captures complex 3D light scattering information from live specimens (2023, August 15)