For years, capturing detailed three-dimensional images of complex biological specimens has posed a major challenge due to their intricate composition and the multiple scattering of light. A game-changing moment has arrived, as scientists from the MIT Laser Biomedical Research Center and the Chinese University of Hong Kong have introduced an innovative method called speckle diffraction tomography (SDT).
In a remarkable feat, the research team harnessed the power of SDT to produce high-resolution images of thick biological samples, achieving an impressive lateral resolution of approximately 500 nanometers and an axial resolution of 1 micrometer using a reflection-based full-field optical setup. The significance of this achievement lies in the capability of SDT to reveal even the tiniest height variations on tissue surfaces, down to the nanometer scale.
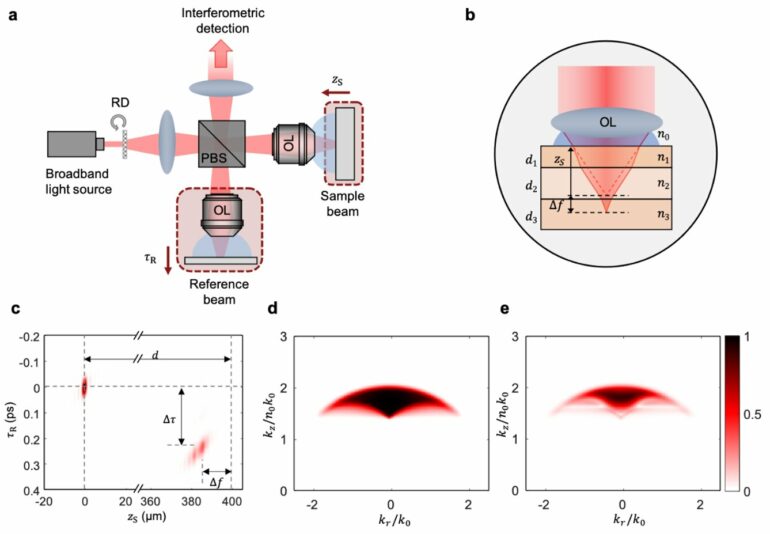
The SDT method leverages dynamic speckle-field interferometry and a low-coherence light source to mitigate the unwanted multiple-scattering and out-of-focus signals, offering remarkable optical sectioning in full-field imaging. Combined with an advanced inverse scattering model that takes specimen-induced distortions into consideration, this unique spatiotemporal gating mechanism facilitates mapping of refractive index distributions in multi-layered tissue specimens.
The power of SDT is highlighted through simulations showcasing its performance, including spatial frequency coverage and resolution at various depths. Moreover, the researchers have developed a 3D deconvolution algorithm that improves spatial resolution by almost 30%, elevating the precision of the technique even further.
In essence, SDT now makes it possible to capture detailed label-free images of thick biological specimens, boasting an unparalleled lateral resolution of approximately 500 nanometers and an axial resolution of around 1 micrometer, all within a reflection-based full-field optical setup. This revolutionary achievement sets the stage for unprecedented 3D label-free in vivo imaging applications that were once deemed unfeasible.
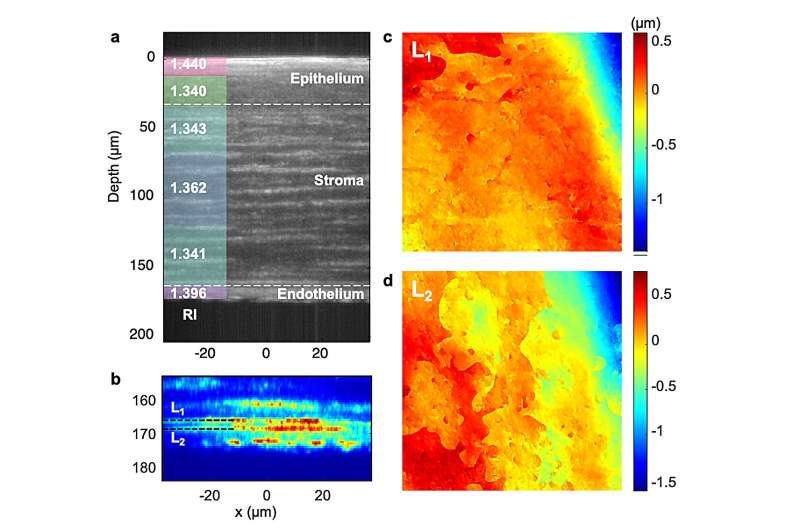
(a) Sectional intensity image of a rat corneal tissue with log scale color mapping (unit in dB). Average RI value of each layer is displayed on the left side. b Magnified image of (a) at depth 155 to 185 µm with linear scale color mapping. Two distinct layers associated with the Dua’s membrane and the Descemet’s membrane are clearly identified and labeled as L1, and L2, respectively. c, d Corresponding height maps of the layers L1 and L2 measured using the SDT system. © by Sungsam Kang, Renjie Zhou, Marten Brelen, Heather K. Mak, Yuechuan Lin, Peter T.C. So and Zahid Yaqoob
The capabilities of SDT were put to the test in imaging red blood cells and quantifying their membrane fluctuations behind a challenging turbid medium, spanning 2.8 scattering mean-free-paths. SDT’s high-resolution and full-field quantitative imaging capabilities were pivotal in enabling this breakthrough.
Additionally, the researchers accomplished volumetric imaging of the cornea within an ex vivo rat eye, shedding light on its optical properties. Notably, SDT unveiled nanoscale topographic features of Dua’s and Descemet’s membranes, an uncharted territory until now.
The results of this research are published in Light: Science & Applications.
More information:
Sungsam Kang et al, Mapping nanoscale topographic features in thick tissues with speckle diffraction tomography, Light: Science & Applications (2023). DOI: 10.1038/s41377-023-01240-0
Provided by
Chinese Academy of Sciences
Citation:
Speckle diffraction tomography reveals nanoscale features in thick biological specimens (2023, August 23)