To better understand how RNA in bacteria gives rise to protein—and along the way, target these processes in the design of new antibiotics—researchers are turning their attention to the unique way this process happens in bacteria.
In eukaryotic cells, transcription (the process by which information in a DNA strand is copied into messenger RNA) and translation (the process by which a protein is synthesized by the ribosome from the mRNA) are two successive steps. In bacteria, they occur simultaneously: As the RNA is being synthesized by RNA polymerase, the ribosome comes in to make the proteins.
This synchronicity allows for so-called “transcription-translation coupling,” wherein the first ribosome can immediately follow and couple with the transcribing RNA polymerase. It is a new area of research that promises to bring insights into processes unique to bacteria that could be targeted with great specificity in the design of antibiotics.
Now, University of Michigan researchers have directly observed previously hidden RNA regulatory mechanisms within such couplings. The results, spearheaded jointly by postdoctoral fellows Surajit Chatterjee and Adrien Chauvier of the U-M Department of Chemistry and the U-M Center for RNA Biomedicine, are published in the Proceedings of the National Academy of Sciences.
The new results promise to have important implications for the future design of antibiotics that could target the coupling mechanism instead of targeting the transcription or translation processes separately.
“With RNA emerging as a major factor in our daily lives—note the SARS-CoV-2 viral genome and the mRNA vaccines to combat its replication—we are at a crossroads where the interplay between RNAs and proteins in their ubiquitous complexes becomes an attractive prospective target for the medicines of the future, including to fight drug-resistant bacterial strains,” said senior author Nils Walter, professor of chemistry.
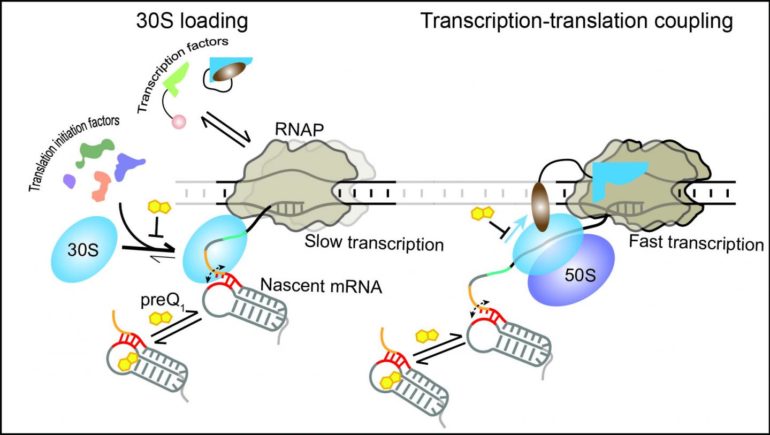
In particular, the researchers found that modulating the translation of a nascent mRNA affects the downstream synthesis of the mRNA itself. When translation is stopped or delayed, the transcription rate is slowed down to avoid overproduction of RNA that would only be degraded in the cell.
To conveniently modulate translation efficiency, the researchers exploited the features of a structured RNA, called a translational riboswitch, embedded near the start of an mRNA of the anthrax bacterium Bacillus anthracis. This RNA changes structure when binding a specific small ligand to reduce translation in response to environmental cues.
The current study shows that the riboswitch—generally thought to only affect translation—can in fact regulate both translation and transcription by exploiting their coupling. By using the riboswitch ligand to slow translation initiation, or inhibitors to delay or stop translation, the scientists observed effects also on the speed of RNA polymerase.
The authors expanded a combination of single-molecule fluorescence microscopy techniques to monitor the dynamic interactions of the transcription and translation machineries during different stages of coupling. They also developed a unique strategy to directly watch transcription-translation coupling in real-time, detecting that the small riboswitch controls the much larger transcription and translation machineries. The work thus surpasses and brings to life previous structure-based studies that provided only snapshots of the already coupled machineries.
The researchers say their results establish important foundations for future RNA research. They explain that the question of how other cellular factors contribute to establishing and maintaining transcription-translation coupling is still enigmatic, raising questions that remain to be investigated. This work could also bring insights into similar biological processes in other pathogenic organisms.
“It is fascinating to see how the huge transcription and translation machineries are held by a tiny mRNA for a controlled gene expression process in bacteria,” said Chatterjee.
Chatterjee and Chauvier are senior postdoctoral fellows in the Walter lab within the U-M Department of Chemistry. They are interested in translational and transcriptional riboswitches, respectively. In this study, they combined their knowledge and interest for each aspect of the coupling.
“To me, it’s not so much about bacteria, but rather about the biological processes of translation and transcription,” Chauvier said. “Genetic regulation is a timely coordinated process and synchronization is the key for the bacteria to adapt to external threats.”
Chatterjee, Chauvier and Walter were joined in the effort by graduate student Shiba Dandpat and collaborator Professor Irina Artsimovitch of Ohio State University.
Synthetic protein quality control system in bacteria
More information:
Surajit Chatterjee et al, A translational riboswitch coordinates nascent transcription–translation coupling, Proceedings of the National Academy of Sciences (2021). DOI: 10.1073/pnas.2023426118
Provided by
University of Michigan
Citation:
RNA holds the reins in bacteria: Researchers observe RNA controlling protein synthesis (2021, April 14)
retrieved 14 April 2021
from https://phys.org/news/2021-04-rna-reins-bacteria-protein-synthesis.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.