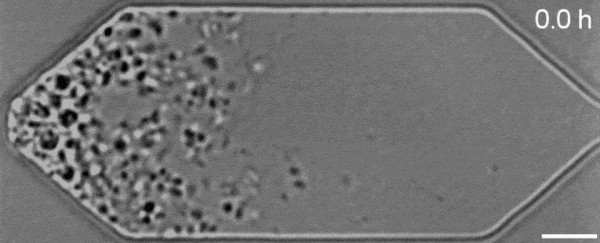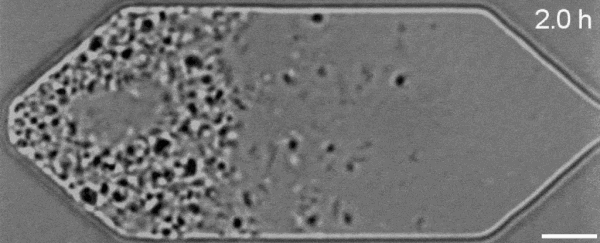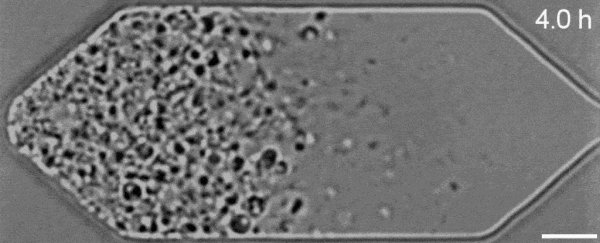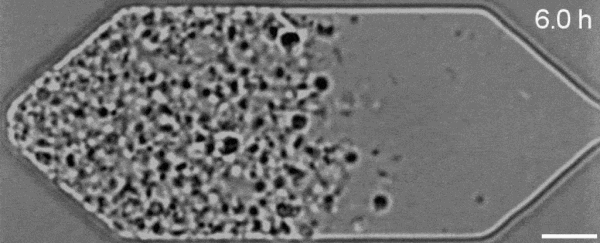
In a new first for genetic engineering, scientists have developed a single-celled synthetic organism that grows and divides much like a normal cell, mimicking aspects of the cell division cycle that underlies and generates healthy living cellular life.
The achievement, demonstrated in an engineered unicellular bacteria-like life form called JCVI-syn3A, is the result of decades of genomic sequencing and analysis by scientists, exploring the roles individual genes play inside living creatures.
“Our goal is to know the function of every gene so we can develop a complete model of how a cell works,” says biophysicist James Pelletier from MIT and the National Institute of Standards and Technology (NIST).
While the work’s roots can be traced back to the 1990s, the most recent strides occurred this century, with researchers in 2003 successfully synthesizing a small virus that infects bacteria.
That led to a new breakthrough in 2010, with scientists at the J. Craig Venter Institute (JCVI) in Maryland designing the first synthetic bacterial cell, called JCVI-syn1.0: the first organism on Earth with an entirely synthetic genome, engineered by stripping the natural DNA out of the bacterium Mycoplasma mycoides.
Several years later, the team took another step forward, creating a species of bacteria in the lab with a genetic code smaller than any found in nature.
This organism, called JCVI-syn3.0, only possessed 473 genes in total – shorter than any known self-sustaining, living organism in the natural world.
But while JCVI-syn3.0’s miniaturized genetic toolkit enabled it to perpetuate itself via cell division, it did so in an unusual way, producing “striking morphological variation” in the new cells it created, which emerged in a variety of different shapes and sizes.
Now, members of the same research team have figured out a way to prevent these strange morphologies from occurring, with a newly modified variant of JCVI-syn3.0, known as JCVI-syn3A.
With the addition of 19 genes not present in JCVI-syn3.0, the newfangled JCVI-syn3A is able to undergo cell division in a more normal-looking, consistent way, with significantly less morphological variation than JCVI-syn3.0 exhibited.
Despite the several years of work behind the achievement, there’s still a huge amount of mystery wrapped up in these genes.
For example, while JCVI-syn3A features 19 new genes, only 7 genes are thought to play a role in making its cell division processes run in a more regular fashion. And of those seven, only two genes – called ftsZ and sepF – have had their functions identified.
Quite how the other five necessarily contribute to JCVI-syn3A’s morphological consistency remains unknown, but one thing is certain: this tiny genome now represents the new standard for experimentation that could help us characterize just what these genes do inside organisms.
“JCVI-syn3A thus offers a compelling minimal model for bacterial physiology and platform for engineering biology broadly,” the researchers explain in their paper.
Or, to put it another way, as the leader of NIST’s Cellular Engineering Group, Elizabeth Strychalski, says: “We want to understand the fundamental design rules of life. If this cell can help us to discover and understand those rules, then we’re off to the races.”
The findings are reported in Cell.