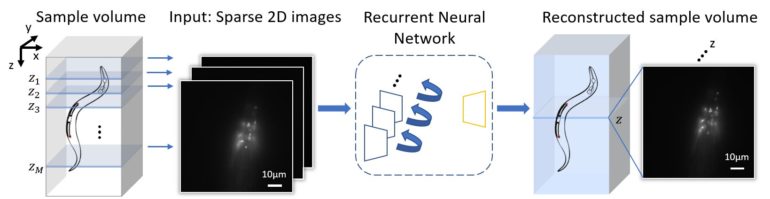
Rapid 3D microscopic imaging of fluorescent samples has numerous applications in physical and biomedical sciences. Given the limited axial range that a single 2D image can provide, 3D fluorescence imaging often requires time-consuming mechanical scanning of samples using a dense sampling grid. In addition to being slow and tedious, this approach also introduces additional light exposure on the sample, which might be toxic and cause unwanted damage, such as photo-bleaching.
By devising a new recurrent neural network, UCLA researchers have demonstrated a deep learning-enabled volumetric microscopy framework for 3D imaging of fluorescent samples. This new method only requires a few 2D images of the sample to be acquired for reconstructing its 3D image, providing ~30-fold reduction in the number of scans required to image a fluorescent volume. The convolutional recurrent neural network that is at the heart of this 3D fluorescence imaging method intuitively mimics the human brain in processing information and storing memories, by consolidating frequently appearing and important object information and features, while forgetting or ignoring some of the redundant information. Using this recurrent neural network scheme, UCLA researchers successfully incorporated spatial features from multiple 2D images of a sample to rapidly reconstruct its 3D fluorescence image.
Published in Light: Science and Applications, the UCLA team demonstrated the success of this volumetric imaging framework using fluorescent C. Elegans samples, which are widely used as a model organism in biology and bioengineering. Compared with standard wide-field volumetric imaging that involves densely scanning of samples, this recurrent neural network-based image reconstruction approach provides a significant reduction in the number of required image scans, which also lowers the total light exposure on the sample. These advances offer much higher imaging speed for observing 3D specimen, while also mitigating photo-bleaching and phototoxicity related challenges that are frequently observed in 3D fluorescence imaging experiments of live samples.
This research is led by Professor Aydogan Ozcan, an associate director of the UCLA California NanoSystems Institute (CNSI) and the Volgenau Chair for Engineering Innovation at the electrical and computer engineering department at UCLA. The other authors include graduate students Luzhe Huang, Hanlong Chen, Yilin Luo and Professor Yair Rivenson, all from electrical and computer engineering department at UCLA. Ozcan also has UCLA faculty appointments in bioengineering and surgery, and is an HHMI professor.
Autofocusing of microscopy images using deep learning
More information:
Luzhe Huang et al. Recurrent neural network-based volumetric fluorescence microscopy, Light: Science & Applications (2021). DOI: 10.1038/s41377-021-00506-9
Provided by
UCLA Engineering Institute for Technology Advancement
Citation:
3D fluorescence microscopy gets a boost using recurrent neural networks (2021, March 23)
retrieved 24 March 2021
from https://techxplore.com/news/2021-03-3d-fluorescence-microscopy-boost-recurrent.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.