Antibiotic resistance is a growing public health problem around the world. When bacteria like E. coli no longer respond to antibiotics, infections become harder to treat.
To develop new antibiotics, researchers typically identify the genes that make bacteria resistant. Through laboratory experiments, they observe how bacteria respond to different antibiotics and look for mutations in the genetic makeup of resistant strains that allow them to survive.
While effective, this method can be time-consuming and may not always capture the full picture of how bacteria become resistant. For example, changes in how genes work that don’t involve mutations can still influence resistance. Bacteria can also exchange resistance genes between each other, which may not be detected if only focusing on mutations within a single strain.
My colleagues and I developed a new approach to identify E. coli resistance genes by computer modeling, allowing us to design new compounds that can block these genes and make existing treatments more effective.
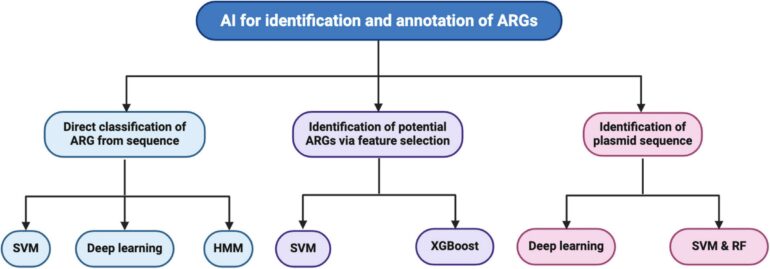
Identifying resistance
To predict which genes contribute to resistance, we analyzed the genomes of various E. coli strains to identify genetic patterns and markers associated with resistance. We then used machine learning algorithms trained on existing data to highlight novel genes or mutations shared across resistant strains that might contribute to resistance.
E. coli is one of many bacterial species developing resistance to common antibiotics.
National Institute of Allergy and Infectious Diseases/National Institutes of Health via Flickr, CC BY-NC
After identifying resistance genes, we designed inhibitors that specifically target and block the proteins these genes produce. By analyzing the structure of the proteins these genes code for, we were able to optimize our inhibitors to strongly bind to these specific proteins.
To reduce the likelihood that bacteria would evolve resistance to these inhibitors, we targeted regions of their genome that code for proteins critical to their survival. By interfering with how bacteria carry out important functions, it makes it more difficult for them to develop mechanisms to compensate. We also prioritized compounds that work differently from existing antibiotics to minimize cross-resistance.
Finally, we tested how effectively our inhibitors could overcome antibiotic resistance in E. coli. We used computer simulations to assess how strongly a number of inhibitors bind to target proteins over time. One inhibitor called hesperidin was able to strongly bind to the three genes in E. coli involved in resistance that we identified, suggesting it may be able to help combat antibiotic-resistant strains.
A global threat
The World Health Organization ranks antimicrobial resistance as one of the top 10 threats to global health. In 2019, bacterial antibiotic resistance killed an estimated 4.95 million people worldwide.
By targeting the…