An interdisciplinary team of biologists and mathematicians at the University of California, Irvine has developed a new tool to help decipher the language cells use to communicate with one another.
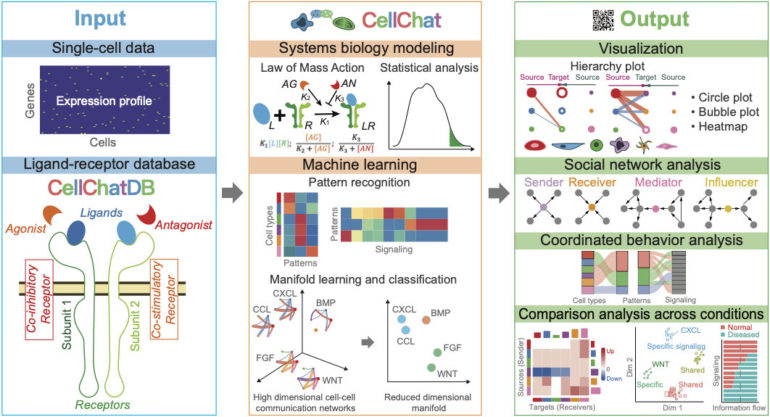
In a paper published today in Nature Communications, the researchers introduce CellChat, a computational platform that enables the decoding of signaling molecules that transmit information and commands between the cells that come together to form biological tissues and even entire organs.
“To properly understand why cells do certain things, and to predict their future actions, we need to be able to listen to what they are saying to one another; mathematical and machine learning tools enable the translation of such messages,” said co-senior author Qing Nie, UCI Chancellor’s Professor of mathematics and developmental & cell biology.
“Just like in our world, where we are constantly bombarded with information, all cells experience a lot of molecular words coming at them simultaneously,” added co-senior author Maksim Plikus, UCI professor of developmental & cell biology, “What they choose to do is dependent on this steady flow of molecular information and on what words and sentences are being heard the loudest.”
To use CellChat to translate molecular messages between cells, researchers feed in a single-cell gene expression, and out comes a detailed report on signaling communication features of a given tissue or organ.
“For each distinct group of cells, CellChat shows what significant signals are being sent to their neighbors and what signals they have the ability to receive,” Plikus said. “As an interpreter of cellular language, CellChat provides scientists with a valuable insight into signaling patterns that guide function of the entire organ.”
In developing CellChat, the researchers in UCI’s NSF-Simons Center for Multiscale Cell Fate Research—including postdoctoral fellows Suoqin Jin, Christian F. Guerrero-Juarez, Raul Ramos and Lihua Zhang—borrowed heavily from machine learning tools and social network theory, which allows the platform to predict a higher level meaning of cellular language and identify contextual similarities that are otherwise not obvious. It breaks down the immense complexity of cellular communication patterns.
Cells produce modifier molecules to add emphasis to a certain command, transforming “do this” to “do this now.” CellChat automatically calibrates the strength of signaling communication between cells by considering all significantly present modifier molecules. As a result, its translation becomes more nuanced and helps to minimize inaccuracies that plague other similar yet less sophisticated computational tools.
Beyond the purely fundamental research enterprise of interpreting these biological messages, Nie said CellChat can also be used to compare communication networks in different states of an organ, such as sickness and health. Calling it a “Google Translator for the lexicon of cells,” Nie said one of tool’s most significant capabilities is that it can be used to uncover molecular drivers in a broad spectrum of maladies including cancer and autoimmune disorders.
“In our paper, we showcase the power of CellChat using atopic dermatitis, a human skin condition, but the tool can be used on any tissue with the same success,” Plikus added.
Researchers use deep learning to identify gene regulation at single-cell level
More information:
Nature Communications (2021). DOI: 10.1038/s41467-021-21246-9
Provided by
University of California, Irvine
Citation:
Researchers eavesdrop on cellular conversations (2021, February 18)
retrieved 19 February 2021
from https://phys.org/news/2021-02-eavesdrop-cellular-conversations.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.