After so many years learning how microbes work, researchers are now digitally recreating their inner workings to tackle challenges ranging from climate change to space colonization.
In my work as a computational biologist, I research ways to get microbes to produce more useful chemicals, such as fuels and bioplastics, that can be used in the energy, agricultural or pharmaceutical industries. Traditionally, researchers have to conduct several trial-and-error experiments on petri dishes in order to determine the optimal conditions microbes need to produce high amounts of chemicals.
Instead, I am able to simulate these experiments all from behind a computer screen through digital blueprints that replicate the inside of microbes. Called genome-scale metabolic models, or GEMs, these virtual labs significantly reduce the time and cost required to figure out what researchers need to do to get what they’re looking for. With GEMs, researchers cannot only explore the complex network of metabolic pathways that allow living organisms to function, but also tweak, test and predict how microbes would behave in different environments, including on other planets.
As GEM technology continues to evolve, I believe these models will play an increasingly important role in shaping the future of biotechnology, medicine and space exploration.
What are genome-scale metabolic models?
Genome-scale metabolic models are digital maps of all the known chemical reactions that occur in cells – that is, the cell’s metabolism. These reactions are crucial for converting food into energy, building cellular structures and detoxifying harmful substances.
To create a GEM, I begin by analyzing an organism’s genome, which contains the genetic instructions cells use to produce proteins. A type of protein coded in the genome called enzymes are the workhorses of metabolism – they facilitate the conversion of nutrients into energy and building blocks for cells.
By linking the genes that encode enzymes to the chemical reactions they help make happen, I can build a comprehensive model that maps out the connections between genes, reactions and metabolites.
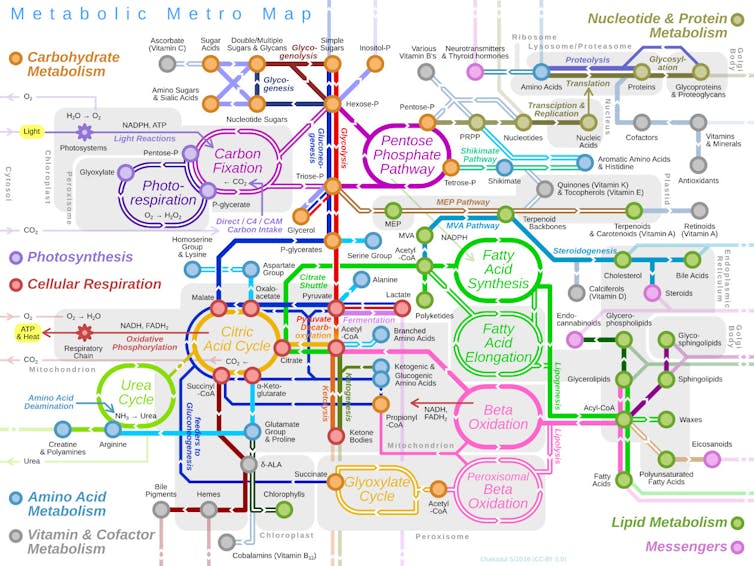
This map shows just a few of the major metabolic pathways in cells.
Chakazul/Wikimedia Commons
Once I build a GEM, I use some advanced computational simulations to make it work like a live cell or microbe would. One of the most common algorithms researchers use to do these simulations is called a flux balance analysis. This mathematical algorithm analyzes available data about metabolism, then makes predictions on how different chemical reactions and metabolites would act under specific conditions.
This makes GEMs particularly useful for understanding how organisms respond to genetic changes and environmental stresses. For example, I can use this method to predict how an organism will react when a specific gene is knocked out. I could also use it to predict how it might…



